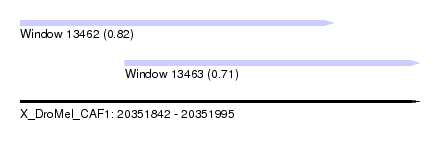
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,351,842 – 20,351,995 |
| Length | 153 |
| Max. P | 0.816478 |

| Location | 20,351,842 – 20,351,962 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -42.26 |
| Consensus MFE | -35.68 |
| Energy contribution | -36.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20351842 120 + 22224390 ACACGAGUCCCUACUUAACCCCCGAGAAUCUCAUCGAGCGCACCGUCGACGUCCUGCUCGCCGAACAUCCUGGCGAGCUGGUCAAGACGGGAUCUCCACAUGUGGUGCGUACUCAUAUCU ....((((..............(((........))).((((((((((...(.((.((((((((.......)))))))).)).)..))).((....))......)))))))))))...... ( -42.60) >DroSec_CAF1 19651 120 + 1 ACACCAGUCCCUACUUGACCCCCGAGAAUCUCAUCGAGCGCACCGUCGACGUCCUGCUCGCCGAACAUCCUGGCGAGCUGGUCAAGACGGGGUCGCCACAUGUGGUGCGUACCCAUAUCU .(((((.........((((((((((........)))........(((...(.((.((((((((.......)))))))).)).)..)))))))))).......)))))............. ( -41.29) >DroSim_CAF1 20764 120 + 1 ACACGAGUCCCUACUUGACCCCCGAGAAUCUCAUCGAGCGCACCGUCGACGUCCUGCUCGCCGAACAUCCUGGCGAGCUGGUCAAGACGGGGUCUCCACAUGUGGUGCGUACCCAUAUCU ....(((.((((.(((((((...(((...))).((((.((...))))))......((((((((.......)))))))).)))))))..)))).)))...((((((.......)))))).. ( -42.30) >DroEre_CAF1 16917 120 + 1 ACACGAGUCCCUACUUGACCCCCGAGAAUCUCAUCGAGCGCACCGUCGACGUCCUGCUCGCCGAGCAUCCUGGCGAGCUGGUCAAGACGGGAUCUCCACAUGUGGUGGGUACUCAUAUCU ....(((((((..((((.....)))).(((.(((.(((..(.(((((...(.((.((((((((.......)))))))).)).)..))))))..)))...))).)))))).))))...... ( -45.10) >DroYak_CAF1 20476 120 + 1 ACACGAGUCCCUACUUGACCCCCGAGAAUCUCAUCGAGCGCACCGUCGACGUCCUGCUAGCCGAACAUCCUGGCGAGCUGGUCAAGACGGGGUCUCCACAUGUGGUGGGUACACCUAUCU ....(((.((((.(((((((..(((........)))((((((.((....))...)))..((((.......))))..))))))))))..)))).))).......((((((....)))))). ( -40.00) >consensus ACACGAGUCCCUACUUGACCCCCGAGAAUCUCAUCGAGCGCACCGUCGACGUCCUGCUCGCCGAACAUCCUGGCGAGCUGGUCAAGACGGGGUCUCCACAUGUGGUGCGUACCCAUAUCU ...(((((....))))).....(((........))).((((((((((...(.((.((((((((.......)))))))).)).)..))).((....))......))))))).......... (-35.68 = -36.68 + 1.00)


| Location | 20,351,882 – 20,351,995 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.26 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -29.28 |
| Energy contribution | -29.29 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20351882 113 + 22224390 CACCGUCGACGUCCUGCUCGCCGAACAUCCUGGCGAGCUGGUCAAGACGGGAUCUCCACAUGUGGUGCGUACUCAUAUCUCA-------UAACUGCAAAGAUUUUAACUUUCAGCAAACU ..(((((...(.((.((((((((.......)))))))).)).)..))))).........(((.((((.(....).)))).))-------)..(((.((((.......)))))))...... ( -33.00) >DroSec_CAF1 19691 113 + 1 CACCGUCGACGUCCUGCUCGCCGAACAUCCUGGCGAGCUGGUCAAGACGGGGUCGCCACAUGUGGUGCGUACCCAUAUCUCA-------UAACUGCAAAGAUUUUUACUUUAAGCAAACU ...((((...(.((.((((((((.......)))))))).)).)..))))(((((((((....)))))...))))........-------....(((((((.......))))..))).... ( -37.70) >DroSim_CAF1 20804 113 + 1 CACCGUCGACGUCCUGCUCGCCGAACAUCCUGGCGAGCUGGUCAAGACGGGGUCUCCACAUGUGGUGCGUACCCAUAUCUCA-------UAGCUGCAAAGAUUUUUACUUUAAGCAAACU ...((((...(.((.((((((((.......)))))))).)).)..))))(((((.(((....)))...).))))........-------..(((..((((.......)))).)))..... ( -36.80) >DroYak_CAF1 20516 103 + 1 CACCGUCGACGUCCUGCUAGCCGAACAUCCUGGCGAGCUGGUCAAGACGGGGUCUCCACAUGUGGUGGGUACACCUAUCUCAUCUCUCAUAGCUGCACAGAUU----------------- ..(((((...(.((.(((.((((.......)))).))).)).)..)))))(((......(((.((((((....)))))).)))........))).........----------------- ( -30.64) >consensus CACCGUCGACGUCCUGCUCGCCGAACAUCCUGGCGAGCUGGUCAAGACGGGGUCUCCACAUGUGGUGCGUACCCAUAUCUCA_______UAACUGCAAAGAUUUUUACUUUAAGCAAACU ..(((((...(.((.((((((((.......)))))))).)).)..)))))..........((..((.........................))..))....................... (-29.28 = -29.29 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:05 2006