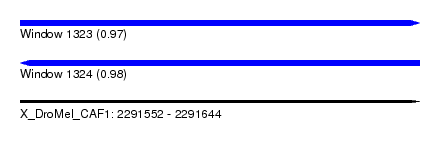
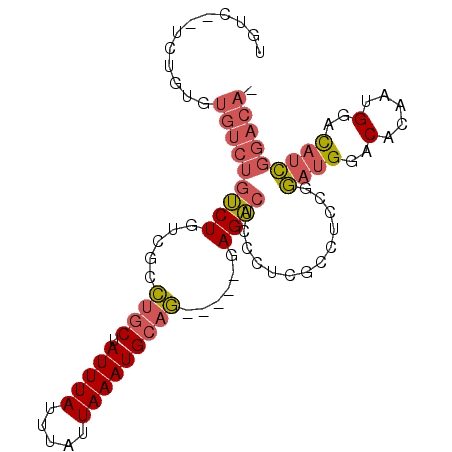
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,291,552 – 2,291,644 |
| Length | 92 |
| Max. P | 0.984447 |

| Location | 2,291,552 – 2,291,644 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -14.75 |
| Energy contribution | -16.83 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2291552 92 + 22224390 -UGUCCGAUGUCCAUUGUGUCCAUCCGGAGGCGAGGGAGUCUC-----CUGCAUUUAAUAAAUAAAUAGCAGGCGACAGACAGACACACAGAUAGACA -((((.....(((.((((.(((....))).)))).)))(((.(-----(((((((((.....))))).))))).))).))))................ ( -30.30) >DroSec_CAF1 5762 92 + 1 -UGUCCGAUGUCCAUUGUGUCCAUCCGGAGGCGAGGGAGUCUC-----CUGCAUUUAAUAAAUAAAUAGCAGGCGACAGACAGACACACAGAGAGACA -.((((..(((((.((((.(((....))).)))).)))(((.(-----(((((((((.....))))).))))).)))...........))..).))). ( -31.50) >DroSim_CAF1 5374 92 + 1 -UGUCCGAUGUCCAUUGUGUCCAUCCGGAGGCGAGGGAGUCUC-----CUGCAUUUAAUAAAUAAAUAGCAGGCGACAGACAGACACACAGAGAGACA -.((((..(((((.((((.(((....))).)))).)))(((.(-----(((((((((.....))))).))))).)))...........))..).))). ( -31.50) >DroEre_CAF1 5753 81 + 1 -UGUCCGAUGUCCAUUGUGUCCAUCCGGAGGCGAGGGAGUCUC-----CUGCAUUUAAUAAAUAAAUAGCAGACGACAGACAGACAG----------- -((((.(...(((.((((.(((....))).)))).)))(((..-----(((((((((.....))))).))))..)))...).)))).----------- ( -26.10) >DroWil_CAF1 18080 81 + 1 -CGACCAAGAUCCAUU--GUCCAUUCUUAUAUAUGGAAGACUUUGGUCUGUCAUUUAAUAAAUAAAUAGCAAGAGACAGC--------------GGCA -.(((((((.((....--.(((((........))))).)).))))))).((((((((.....))))).((........))--------------))). ( -17.90) >DroYak_CAF1 5783 91 + 1 UUGUCCGCUGUCCAUUGUGUCCAUCCGCAGGCGAGGGAGUCUC-----CUGCAUUUAAUAAAUAAAUAGCAGGCGACAGACAGACAGACAGCA--UUU ......((((((...(.((((..(((........))).(((.(-----(((((((((.....))))).))))).))).)))).)..)))))).--... ( -31.30) >consensus _UGUCCGAUGUCCAUUGUGUCCAUCCGGAGGCGAGGGAGUCUC_____CUGCAUUUAAUAAAUAAAUAGCAGGCGACAGACAGACACACAGA__GACA .((((.....(((.((((.(((....))).)))).)))((((......(((((((((.....))))).)))).....)))).))))............ (-14.75 = -16.83 + 2.09)

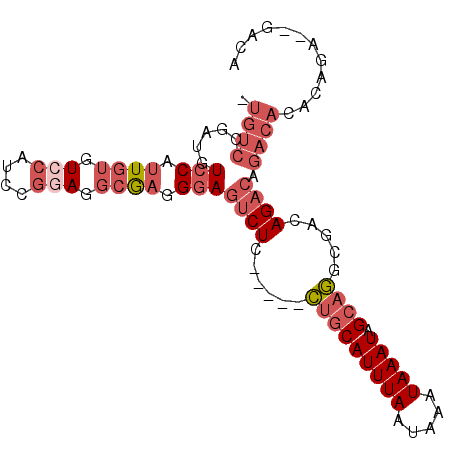
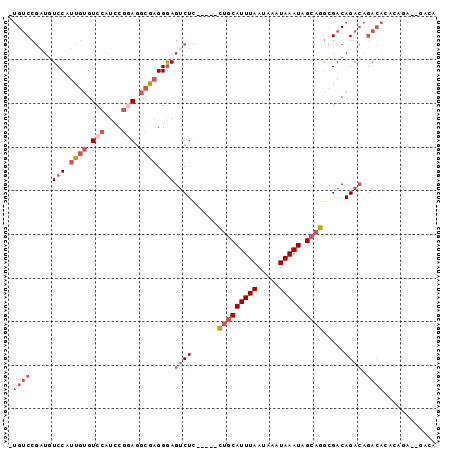
| Location | 2,291,552 – 2,291,644 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2291552 92 - 22224390 UGUCUAUCUGUGUGUCUGUCUGUCGCCUGCUAUUUAUUUAUUAAAUGCAG-----GAGACUCCCUCGCCUCCGGAUGGACACAAUGGACAUCGGACA- (((((((...((((((((((((..((((((.(((((.....)))))))))-----(((.....)))))...))))))))))))))))))).......- ( -33.80) >DroSec_CAF1 5762 92 - 1 UGUCUCUCUGUGUGUCUGUCUGUCGCCUGCUAUUUAUUUAUUAAAUGCAG-----GAGACUCCCUCGCCUCCGGAUGGACACAAUGGACAUCGGACA- .((((.((((((((((((((((..((((((.(((((.....)))))))))-----(((.....)))))...))))))))))).)))))....)))).- ( -32.50) >DroSim_CAF1 5374 92 - 1 UGUCUCUCUGUGUGUCUGUCUGUCGCCUGCUAUUUAUUUAUUAAAUGCAG-----GAGACUCCCUCGCCUCCGGAUGGACACAAUGGACAUCGGACA- .((((.((((((((((((((((..((((((.(((((.....)))))))))-----(((.....)))))...))))))))))).)))))....)))).- ( -32.50) >DroEre_CAF1 5753 81 - 1 -----------CUGUCUGUCUGUCGUCUGCUAUUUAUUUAUUAAAUGCAG-----GAGACUCCCUCGCCUCCGGAUGGACACAAUGGACAUCGGACA- -----------......(((((((..((((.(((((.....)))))))))-----..))).............((((..(.....)..)))))))).- ( -23.10) >DroWil_CAF1 18080 81 - 1 UGCC--------------GCUGUCUCUUGCUAUUUAUUUAUUAAAUGACAGACCAAAGUCUUCCAUAUAUAAGAAUGGAC--AAUGGAUCUUGGUCG- ....--------------..((((..(((.((......)).)))..))))((((((..((((((((........))))).--...)))..)))))).- ( -19.60) >DroYak_CAF1 5783 91 - 1 AAA--UGCUGUCUGUCUGUCUGUCGCCUGCUAUUUAUUUAUUAAAUGCAG-----GAGACUCCCUCGCCUGCGGAUGGACACAAUGGACAGCGGACAA ...--...((((((.(((((((((.(((((.(((((.....)))))))))-----).)))(((.(((....)))..)))......)))))))))))). ( -35.20) >consensus UGUC__UCUGUGUGUCUGUCUGUCGCCUGCUAUUUAUUUAUUAAAUGCAG_____GAGACUCCCUCGCCUCCGGAUGGACACAAUGGACAUCGGACA_ ............(((((((((.....((((.(((((.....)))))))))......)))).............((((..(.....)..))))))))). (-14.00 = -14.70 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:32 2006