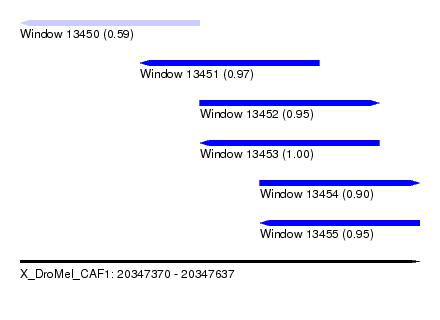
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,347,370 – 20,347,637 |
| Length | 267 |
| Max. P | 0.999683 |

| Location | 20,347,370 – 20,347,490 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -28.24 |
| Energy contribution | -28.04 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20347370 120 - 22224390 ACCCGAAGAGUGAGUGAAAAAUCGCAACCACAUAGGGCACGGCCGCCCGCCCAUCGACGAAAAUAUACAAAAAAAAUUUCCUACAGGGCGGUGCUAAAUACUUGCUUGGGUUUUUCGGCU ((((((.(((((.((((....)))).(((.....((((......))))((((......((((..............)))).....)))))))......)))))..))))))......... ( -32.44) >DroSec_CAF1 15217 118 - 1 ACCCGAAGAGUGAGUAAAAAUUCGCAACCACAUAGGGCACGGCCGCCCGCCCAUCGACGAAAAUAUACAAA--AAAUUCCCUACAGGGCGGUGCUAAAUACUUGCUUAGGUUUUUCGGCU ..(((((((((((((....)))))).(((......(((...)))((((((((......(((..........--...)))......)))))).))..............)))))))))).. ( -32.22) >DroSim_CAF1 16981 118 - 1 ACCCGAAGAGUGAGUGAAAAAUCGCAACCACAUAGGGCACGGCCGCCCGCCCAUCGACGAAAAUAUACAAA--AAAUUCCCUACAGGGCGGUGCUAAAUACUUGCUUGGGUUUUUCGGCU ((((((.(((((.((((....)))).(((.....((((......))))((((......(((..........--...)))......)))))))......)))))..))))))......... ( -32.02) >DroEre_CAF1 11976 120 - 1 ACCCGAAGAAUGAGUGAAAAAUCGCAACCACAUAGGGCACAGCCGUCCGCCCAUCGACGAAAAUGUACAAAAAAAAUUCCCUACAGGGCGGUGCUAAAUACUUCCUCCGGUUUUUCGGUU ..((((((((((((.(((.....(((.((.....((((......))))((((.((...))...((((..............))))))))))))).......))))))..))))))))).. ( -29.74) >DroYak_CAF1 15802 119 - 1 AGCCGAAGAAUGAGUGAAAAAUCGCAACCACACAGGGCACAGUCGCCCGCCCACCGAUGAAAAUAUACAAAA-AAAUUCCCUACAGGGCGGUGCUAAAUACUUCCUUGGGUUUUACGGUU (((((.(((((..((((....))))..(((....((((......))))...(((((((....))........-.....(((....)))))))).............)))))))).))))) ( -30.70) >consensus ACCCGAAGAGUGAGUGAAAAAUCGCAACCACAUAGGGCACGGCCGCCCGCCCAUCGACGAAAAUAUACAAAA_AAAUUCCCUACAGGGCGGUGCUAAAUACUUGCUUGGGUUUUUCGGCU ..(((((((((((((((......(((.((.....((((......))))((((......(((...............)))......))))))))).......)))))))..)))))))).. (-28.24 = -28.04 + -0.20)



| Location | 20,347,450 – 20,347,570 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -40.14 |
| Consensus MFE | -34.00 |
| Energy contribution | -34.16 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20347450 120 - 22224390 AAGCCAGGGCGAAAGAGAUGGCAUCUGCGUGCGAGAAAGAGAGCGCCAAUGUCUUGUGGCUGCCAUUUGCGGCGAAGAAAACCCGAAGAGUGAGUGAAAAAUCGCAACCACAUAGGGCAC ..((((...(....)...))))...((.((((..........)))))).((((((((((((((.....)))))................((((.(....).)))).....))))))))). ( -36.60) >DroSec_CAF1 15295 120 - 1 AAGCCAGGGCGAAAGAGAUGGCCUCUGCGCGCGAGAAAGAGAGCGCCAAUGUCUUGUGGCUGCCAUUUGCGGCCAAGAAAACCCGAAGAGUGAGUAAAAAUUCGCAACCACAUAGGGCAC ..(((.(((...((((.(((((((((.(.(....)...))))).))).)).)))).(((((((.....)))))))......))).....((((((....))))))..........))).. ( -42.70) >DroSim_CAF1 17059 120 - 1 AAGCCAGGGCGAAAGAGAUGGCCUCUGCGCGCGAGAAAGAGAGCGCCAAUGUCUUGUGGCUGCCAUUUGCGGCCAAGAAAACCCGAAGAGUGAGUGAAAAAUCGCAACCACAUAGGGCAC ..(((.(((...((((.(((((((((.(.(....)...))))).))).)).)))).(((((((.....)))))))......))).....(((.((((....))))...)))....))).. ( -42.10) >DroEre_CAF1 12056 120 - 1 AAGCCAGGGCGAAAGAGAUGGCCUCUGCGUGGGCGAAAGAGAGCGCCGAUGUCUUGUGGCUGCCAUUUGCGGCCAAGAAAACCCGAAGAAUGAGUGAAAAAUCGCAACCACAUAGGGCAC ..(((.(((...((((.(((((((((.((....))....)))).))).)).)))).(((((((.....)))))))......))).........((((....))))..........))).. ( -39.40) >DroYak_CAF1 15881 120 - 1 AAGCCAGGGCGAAAGAGACGGCCUCUGCGUGCGAGAAAGAGAGCGCCGAUGUCUUGUGGCUGCCAUUUGCGGCCAAGAAAAGCCGAAGAAUGAGUGAAAAAUCGCAACCACACAGGGCAC ..(((..(((..((((.(((((((((.(..........))))).)))).).)))).(((((((.....)))))))......))).........((((....))))..........))).. ( -39.90) >consensus AAGCCAGGGCGAAAGAGAUGGCCUCUGCGUGCGAGAAAGAGAGCGCCAAUGUCUUGUGGCUGCCAUUUGCGGCCAAGAAAACCCGAAGAGUGAGUGAAAAAUCGCAACCACAUAGGGCAC ..(((.(((...((((.(((((((((.(.(....)...))))).)))).).)))).(((((((.....)))))))......))).........((((....))))..........))).. (-34.00 = -34.16 + 0.16)

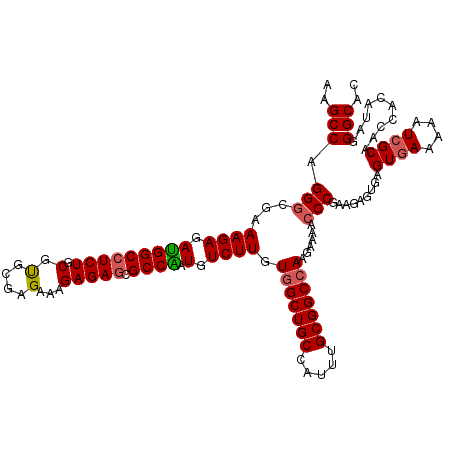
| Location | 20,347,490 – 20,347,610 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -31.62 |
| Energy contribution | -31.98 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20347490 120 + 22224390 UUUCUUCGCCGCAAAUGGCAGCCACAAGACAUUGGCGCUCUCUUUCUCGCACGCAGAUGCCAUCUCUUUCGCCCUGGCUUACCAAUUCCUGGUUUGCCACAAAGCCGCGUCUCCAACGCU ......(((.((..((((((((((........))))((..........)).......))))))...........((((..((((.....))))..))))....)).)))........... ( -31.40) >DroSec_CAF1 15335 120 + 1 UUUCUUGGCCGCAAAUGGCAGCCACAAGACAUUGGCGCUCUCUUUCUCGCGCGCAGAGGCCAUCUCUUUCGCCCUGGCUUACCAAUUCCUGGUUUGCCACAAAGCCGCGUCUCCAACGCU ..(((((((.((.....)).))))..)))....(((..............(((.((((.....))))..)))..((((..((((.....))))..))))....)))((((.....)))). ( -35.50) >DroSim_CAF1 17099 120 + 1 UUUCUUGGCCGCAAAUGGCAGCCACAAGACAUUGGCGCUCUCUUUCUCGCGCGCAGAGGCCAUCUCUUUCGCCCUGGCUUACCAAUUCCUGGUUUGCCACAAAGCCGCGUCUCCAACGCU ..(((((((.((.....)).))))..)))....(((..............(((.((((.....))))..)))..((((..((((.....))))..))))....)))((((.....)))). ( -35.50) >DroEre_CAF1 12096 120 + 1 UUUCUUGGCCGCAAAUGGCAGCCACAAGACAUCGGCGCUCUCUUUCGCCCACGCAGAGGCCAUCUCUUUCGCCCUGGCUUACCAAUUCCUGGUUUGCCACAAAGCCGCGUCUCCAACGCU .....((((.((.....)).))))..((((..((((((.((((...((....))))))))..............((((..((((.....))))..))))....)))).))))........ ( -35.30) >DroYak_CAF1 15921 120 + 1 UUUCUUGGCCGCAAAUGGCAGCCACAAGACAUCGGCGCUCUCUUUCUCGCACGCAGAGGCCGUCUCUUUCGCCCUGGCUUACCAAUUCCUGGUUUGCCACAAAGCCGCGUCUCCAACGCU .....((((.((.....)).)))).((((...((((.(((((..........).))))))))..))))..((..((((..((((.....))))..))))....)).((((.....)))). ( -35.40) >consensus UUUCUUGGCCGCAAAUGGCAGCCACAAGACAUUGGCGCUCUCUUUCUCGCACGCAGAGGCCAUCUCUUUCGCCCUGGCUUACCAAUUCCUGGUUUGCCACAAAGCCGCGUCUCCAACGCU .....((((.((.....)).)))).((((...((((.(((((..........).))))))))..))))..((..((((..((((.....))))..))))....)).((((.....)))). (-31.62 = -31.98 + 0.36)


| Location | 20,347,490 – 20,347,610 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -46.64 |
| Consensus MFE | -44.04 |
| Energy contribution | -44.16 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20347490 120 - 22224390 AGCGUUGGAGACGCGGCUUUGUGGCAAACCAGGAAUUGGUAAGCCAGGGCGAAAGAGAUGGCAUCUGCGUGCGAGAAAGAGAGCGCCAAUGUCUUGUGGCUGCCAUUUGCGGCGAAGAAA ...........(((.((...((((((..((((((((((((..((((...(....)...))))....((...(......)...)))))))).))))).)..))))))..)).)))...... ( -44.40) >DroSec_CAF1 15335 120 - 1 AGCGUUGGAGACGCGGCUUUGUGGCAAACCAGGAAUUGGUAAGCCAGGGCGAAAGAGAUGGCCUCUGCGCGCGAGAAAGAGAGCGCCAAUGUCUUGUGGCUGCCAUUUGCGGCCAAGAAA .(((((...)))))((((..((((((..((((((((((((..((((...(....)...))))((((...(....)..))))...)))))).))))).)..))))))....))))...... ( -49.20) >DroSim_CAF1 17099 120 - 1 AGCGUUGGAGACGCGGCUUUGUGGCAAACCAGGAAUUGGUAAGCCAGGGCGAAAGAGAUGGCCUCUGCGCGCGAGAAAGAGAGCGCCAAUGUCUUGUGGCUGCCAUUUGCGGCCAAGAAA .(((((...)))))((((..((((((..((((((((((((..((((...(....)...))))((((...(....)..))))...)))))).))))).)..))))))....))))...... ( -49.20) >DroEre_CAF1 12096 120 - 1 AGCGUUGGAGACGCGGCUUUGUGGCAAACCAGGAAUUGGUAAGCCAGGGCGAAAGAGAUGGCCUCUGCGUGGGCGAAAGAGAGCGCCGAUGUCUUGUGGCUGCCAUUUGCGGCCAAGAAA .(((((...)))))((((..((((((..((((((((((((..((((...(....)...))))((((.((....))..))))...)))))).))))).)..))))))....))))...... ( -47.10) >DroYak_CAF1 15921 120 - 1 AGCGUUGGAGACGCGGCUUUGUGGCAAACCAGGAAUUGGUAAGCCAGGGCGAAAGAGACGGCCUCUGCGUGCGAGAAAGAGAGCGCCGAUGUCUUGUGGCUGCCAUUUGCGGCCAAGAAA .(((((...))))).((((..((((..(((((...)))))..))))))))..((((.(((((((((.(..........))))).)))).).)))).(((((((.....)))))))..... ( -43.30) >consensus AGCGUUGGAGACGCGGCUUUGUGGCAAACCAGGAAUUGGUAAGCCAGGGCGAAAGAGAUGGCCUCUGCGUGCGAGAAAGAGAGCGCCAAUGUCUUGUGGCUGCCAUUUGCGGCCAAGAAA .(((((...)))))((((..((((((..((((((((((((..((((...(....)...))))((((...(....)..))))...)))))).))))).)..))))))....))))...... (-44.04 = -44.16 + 0.12)



| Location | 20,347,530 – 20,347,637 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -23.08 |
| Energy contribution | -23.07 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.904105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20347530 107 + 22224390 UCUUUCUCGCACGCAGAUGCCAUCUCUUUCGCCCUGGCUUACCAAUUCCUGGUUUGCCACAAAGCCGCGUCUCCAACGCUUUGCUUUCGCUUGGAUUUUUCUUCUUU ........(.((((((((...)))).....((..((((..((((.....))))..))))....)).)))))(((((.((.........)))))))............ ( -22.20) >DroSec_CAF1 15375 107 + 1 UCUUUCUCGCGCGCAGAGGCCAUCUCUUUCGCCCUGGCUUACCAAUUCCUGGUUUGCCACAAAGCCGCGUCUCCAACGCUUUGCUUUCGCUUGGAUUUUUCUUCGUU .......((((.((((.(((.(......).)))))(((..((((.....))))..))).....))))))..(((((.((.........)))))))............ ( -28.70) >DroSim_CAF1 17139 107 + 1 UCUUUCUCGCGCGCAGAGGCCAUCUCUUUCGCCCUGGCUUACCAAUUCCUGGUUUGCCACAAAGCCGCGUCUCCAACGCUUUGCUUUCGCUUGGAUUUUUCUUCGUU .......((((.((((.(((.(......).)))))(((..((((.....))))..))).....))))))..(((((.((.........)))))))............ ( -28.70) >DroEre_CAF1 12136 107 + 1 UCUUUCGCCCACGCAGAGGCCAUCUCUUUCGCCCUGGCUUACCAAUUCCUGGUUUGCCACAAAGCCGCGUCUCCAACGCUUUGCUUUUGCUUGCAUUUUUCUUCUUU ......((....((((((((..........((..((((..((((.....))))..))))....)).((((.....))))...))))))))..))............. ( -28.00) >consensus UCUUUCUCGCACGCAGAGGCCAUCUCUUUCGCCCUGGCUUACCAAUUCCUGGUUUGCCACAAAGCCGCGUCUCCAACGCUUUGCUUUCGCUUGGAUUUUUCUUCGUU .........((.((.(((((..........((..((((..((((.....))))..))))....)).((((.....))))...))))).)).)).............. (-23.08 = -23.07 + -0.00)


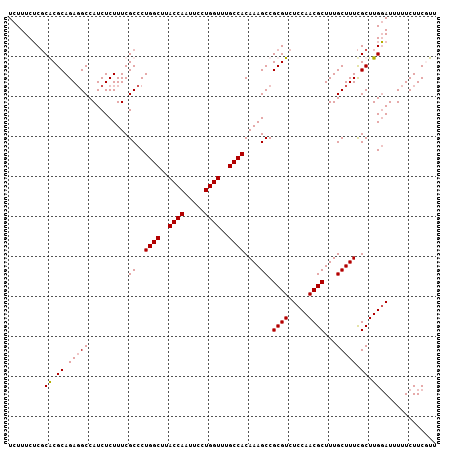
| Location | 20,347,530 – 20,347,637 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -28.54 |
| Energy contribution | -28.35 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20347530 107 - 22224390 AAAGAAGAAAAAUCCAAGCGAAAGCAAAGCGUUGGAGACGCGGCUUUGUGGCAAACCAGGAAUUGGUAAGCCAGGGCGAAAGAGAUGGCAUCUGCGUGCGAGAAAGA .............((..((....))...(((((...)))))))((((.(.(((.(((((...)))))..((((...(....)...)))).......))).).)))). ( -31.40) >DroSec_CAF1 15375 107 - 1 AACGAAGAAAAAUCCAAGCGAAAGCAAAGCGUUGGAGACGCGGCUUUGUGGCAAACCAGGAAUUGGUAAGCCAGGGCGAAAGAGAUGGCCUCUGCGCGCGAGAAAGA .((((((..........((....))...(((((...)))))..))))))(((..(((((...)))))..))).(.(((..((((.....)))).))).)........ ( -32.80) >DroSim_CAF1 17139 107 - 1 AACGAAGAAAAAUCCAAGCGAAAGCAAAGCGUUGGAGACGCGGCUUUGUGGCAAACCAGGAAUUGGUAAGCCAGGGCGAAAGAGAUGGCCUCUGCGCGCGAGAAAGA .((((((..........((....))...(((((...)))))..))))))(((..(((((...)))))..))).(.(((..((((.....)))).))).)........ ( -32.80) >DroEre_CAF1 12136 107 - 1 AAAGAAGAAAAAUGCAAGCAAAAGCAAAGCGUUGGAGACGCGGCUUUGUGGCAAACCAGGAAUUGGUAAGCCAGGGCGAAAGAGAUGGCCUCUGCGUGGGCGAAAGA ............(((..((....(((((((..((....))..))))))).(((.(((((...)))))..((((...(....)...))))...)))))..)))..... ( -31.20) >consensus AAAGAAGAAAAAUCCAAGCGAAAGCAAAGCGUUGGAGACGCGGCUUUGUGGCAAACCAGGAAUUGGUAAGCCAGGGCGAAAGAGAUGGCCUCUGCGCGCGAGAAAGA .................(((...(((((((..((....))..))))))).(((.(((((...)))))..((((...(....)...))))...))).)))........ (-28.54 = -28.35 + -0.19)

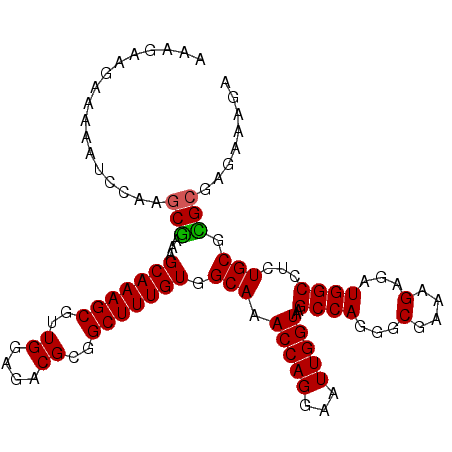
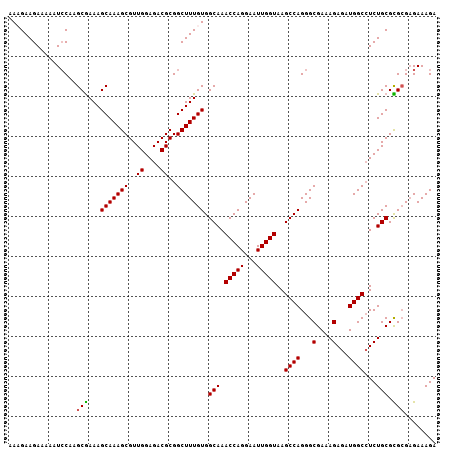
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:57 2006