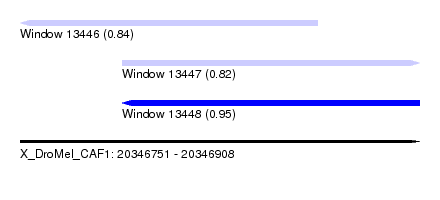
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,346,751 – 20,346,908 |
| Length | 157 |
| Max. P | 0.950143 |

| Location | 20,346,751 – 20,346,868 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.14 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -25.12 |
| Energy contribution | -26.24 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20346751 117 - 22224390 UGGCAAAUUCACCAGCCAGCGGCGGGAAAAUCCCGCUGGAGCCAAGGGAAAAUGGUGGAAAACAGCGAAAUAUC---GCGAAAUAUAACAUAAUAAGAGACAUUUUGAGCCGUUACUAAU .(((...(((((((.((.((((((((.....))))))...))....))....))))))).....((((....))---)).............................)))......... ( -33.60) >DroSec_CAF1 14572 120 - 1 UGGCAAAUUCGCCAGCCAGCGGCGGGAAAAUCCAGCUAGAGCCACGGGAAAAUGGUGGAAAACGGCGAAAAAUCCGGGCCGGAGAAAACAUAAUAAGGGACACUUUGAGCCGUUACUAAU .......(((((((.((.(.(((.((.....)).......))).).))....))))))).((((((......((((...))))..........(((((....))))).))))))...... ( -32.60) >DroSim_CAF1 16357 120 - 1 UGGCAAAUUCGCCAGCCAGCGGCGGGAAAAUCCCGCUGGAGCCACGGGAAAGUGGGGGAAAACGGCGAAAAAUCCGGGCCGGAGAAAACAUAAUAAGGGACACUUGGAGCCGUUACUAAU .(((...((((((..((..(((((((.....)))))))...((((......)))).)).....))))))...((((((((................))....)))))))))......... ( -41.49) >DroEre_CAF1 11370 116 - 1 UGGCAAAUUCGCCAGCCAGCGGCGGGAAAAUCCCGCUGGAGCCACGGGAAAAUGGUGGAAAACAGCGAAAAAGCAUCG--GAAAG--AUAUAAUACGGUACACUUUGAGCCGUUACUAAU ((((......))))...(((((((((.....)))((((...((((.(.....).))))....))))...((((.((((--.....--........))))...))))..))))))...... ( -34.82) >DroYak_CAF1 15162 118 - 1 UGGCAAAUUCGCCAGCCAGCGGCGGGAAAAUCCCGCUGGAGCCGCGGGAAAAUGGUGGAAAACUGCGAAAAAUCAAAG--GAAAAAAACAUAAGUGGAGGCACUUUGAUCCGUUUCUAAU ..(((..(((((((.((.((((((((.....))))))(....))).))....)))))))....)))((((.(((((((--.......((....)).......)))))))...)))).... ( -36.24) >consensus UGGCAAAUUCGCCAGCCAGCGGCGGGAAAAUCCCGCUGGAGCCACGGGAAAAUGGUGGAAAACAGCGAAAAAUCAGGGC_GAAAAAAACAUAAUAAGGGACACUUUGAGCCGUUACUAAU ((((......))))...(((((((((.....)))((((...((((.(.....).))))....))))..........................................))))))...... (-25.12 = -26.24 + 1.12)

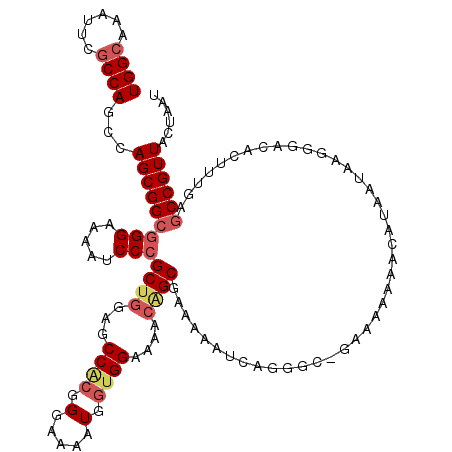

| Location | 20,346,791 – 20,346,908 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.65 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -32.22 |
| Energy contribution | -32.10 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20346791 117 + 22224390 CGC---GAUAUUUCGCUGUUUUCCACCAUUUUCCCUUGGCUCCAGCGGGAUUUUCCCGCCGCUGGCUGGUGAAUUUGCCAGCGACUUGGCAAUCGAAAUCCGCUCAUUGGCUGCAUCGAC .((---((....)))).........(((........))).....(((((.....))))).((.(((..((((..(((((((....))))))).((.....)).))))..)))))...... ( -36.70) >DroSec_CAF1 14612 120 + 1 GGCCCGGAUUUUUCGCCGUUUUCCACCAUUUUCCCGUGGCUCUAGCUGGAUUUUCCCGCCGCUGGCUGGCGAAUUUGCCAGCGACUUGGCAAUCGAAAUCCGCUCAUUGGCUGCAUCGAC (((((((((((((((((.....((((.........))))..((((((((......)))..)))))..)))))..(((((((....)))))))..))))))))......))))........ ( -37.80) >DroSim_CAF1 16397 120 + 1 GGCCCGGAUUUUUCGCCGUUUUCCCCCACUUUCCCGUGGCUCCAGCGGGAUUUUCCCGCCGCUGGCUGGCGAAUUUGCCAGCGCCUUGGCAAUCGAAAUCCGCUCAUUGGCUGCAUCGAC (((((((((((((((((........((((......))))..((((((((.......).)))))))..)))))..(((((((....)))))))..))))))))......))))........ ( -45.10) >DroEre_CAF1 11408 118 + 1 --CGAUGCUUUUUCGCUGUUUUCCACCAUUUUCCCGUGGCUCCAGCGGGAUUUUCCCGCCGCUGGCUGGCGAAUUUGCCAGCGACUUGGCAAUCGAAAUCCGCUCAUUGGCUGCAUCGAC --((((((.....(((((....((((.........))))...)))))(((((((...((((.(.(((((((....))))))).)..))))....)))))))(((....))).)))))).. ( -44.90) >DroYak_CAF1 15202 118 + 1 --CUUUGAUUUUUCGCAGUUUUCCACCAUUUUCCCGCGGCUCCAGCGGGAUUUUCCCGCCGCUGGCUGGCGAAUUUGCCAGCGACUUGGCAAUCGAAAUCCGCUCAUUGGCUGCAUCGAC --............((((((...........((((((.......)))))).......((((.(.(((((((....))))))).)..))))..................))))))...... ( -37.00) >consensus _GCCCGGAUUUUUCGCCGUUUUCCACCAUUUUCCCGUGGCUCCAGCGGGAUUUUCCCGCCGCUGGCUGGCGAAUUUGCCAGCGACUUGGCAAUCGAAAUCCGCUCAUUGGCUGCAUCGAC ...................................((((((..(((.(((((((...((((...(((((((....)))))))....))))....))))))))))....))))))...... (-32.22 = -32.10 + -0.12)


| Location | 20,346,791 – 20,346,908 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.65 |
| Mean single sequence MFE | -43.82 |
| Consensus MFE | -34.78 |
| Energy contribution | -35.70 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20346791 117 - 22224390 GUCGAUGCAGCCAAUGAGCGGAUUUCGAUUGCCAAGUCGCUGGCAAAUUCACCAGCCAGCGGCGGGAAAAUCCCGCUGGAGCCAAGGGAAAAUGGUGGAAAACAGCGAAAUAUC---GCG .................((((((((((.((((((......)))))).(((((((.((.((((((((.....))))))...))....))....)))))))......)))))).))---)). ( -41.70) >DroSec_CAF1 14612 120 - 1 GUCGAUGCAGCCAAUGAGCGGAUUUCGAUUGCCAAGUCGCUGGCAAAUUCGCCAGCCAGCGGCGGGAAAAUCCAGCUAGAGCCACGGGAAAAUGGUGGAAAACGGCGAAAAAUCCGGGCC .........(((......(((((((...(((((..(((((((((..........))))))))).((.....))........((((.(.....).)))).....))))).)))))))))). ( -40.90) >DroSim_CAF1 16397 120 - 1 GUCGAUGCAGCCAAUGAGCGGAUUUCGAUUGCCAAGGCGCUGGCAAAUUCGCCAGCCAGCGGCGGGAAAAUCCCGCUGGAGCCACGGGAAAGUGGGGGAAAACGGCGAAAAAUCCGGGCC .........(((......(((((((...(((((..(((((((((......))))))...(((((((.....)))))))..)))........((........))))))).)))))))))). ( -48.20) >DroEre_CAF1 11408 118 - 1 GUCGAUGCAGCCAAUGAGCGGAUUUCGAUUGCCAAGUCGCUGGCAAAUUCGCCAGCCAGCGGCGGGAAAAUCCCGCUGGAGCCACGGGAAAAUGGUGGAAAACAGCGAAAAAGCAUCG-- ..((((((.((......))((((((...(((((.....((((((......))))))....)))))..))))))(((((...((((.(.....).))))....))))).....))))))-- ( -47.30) >DroYak_CAF1 15202 118 - 1 GUCGAUGCAGCCAAUGAGCGGAUUUCGAUUGCCAAGUCGCUGGCAAAUUCGCCAGCCAGCGGCGGGAAAAUCCCGCUGGAGCCGCGGGAAAAUGGUGGAAAACUGCGAAAAAUCAAAG-- ...(((.((.....)).((((.((((...(((((.(((((((((..........))))))))).......((((((.(....)))))))...))))))))).)))).....)))....-- ( -41.00) >consensus GUCGAUGCAGCCAAUGAGCGGAUUUCGAUUGCCAAGUCGCUGGCAAAUUCGCCAGCCAGCGGCGGGAAAAUCCCGCUGGAGCCACGGGAAAAUGGUGGAAAACAGCGAAAAAUCAGGGC_ (((((..(.((......)).)...)))))......(((((((((..........)))))))))(((.....)))((((...((((.(.....).))))....)))).............. (-34.78 = -35.70 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:51 2006