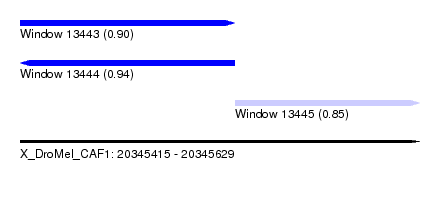
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,345,415 – 20,345,629 |
| Length | 214 |
| Max. P | 0.937459 |

| Location | 20,345,415 – 20,345,530 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -28.30 |
| Energy contribution | -28.55 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20345415 115 + 22224390 --CCACCCACCCACUUCAGUGGCACUUGCCACCGCUUUGCUGCCAUUGUCGUUCCACUUUAGAAUUAUUUGCACAAAAUCUUUCGGCGGCAGCAAGGAUUUCUCGCUACUGUGCUUC --.........(((....(((((....)))))..((((((((((...((((.....)....(((..((((.....))))..)))))))))))))))).............))).... ( -29.60) >DroSec_CAF1 13210 117 + 1 ACCCACCCACCCACUUCAGUGGCACUUGCCACCGCUUUGCUGCCAUUGUCGUUUCACUUUAGAAUUAUUUGCACAAAAUCUUUCGGCGGCAGCAAGGAUUUCUCGCUACUGUGCUUC ...........(((....(((((....)))))..((((((((((...((((.....)....(((..((((.....))))..)))))))))))))))).............))).... ( -29.60) >DroSim_CAF1 14998 116 + 1 AC-CACCCACCCACUUCAGUGGCACUUGCCACCGCUUUGCUGCCAUUGUCGUUCCACUUUAGAAUUAUUUGCACAAAAUCUUUCGGCGGCAGCAAGGAUUUCUCGCUACUGUGCUUC ..-........(((....(((((....)))))..((((((((((...((((.....)....(((..((((.....))))..)))))))))))))))).............))).... ( -29.60) >DroEre_CAF1 9932 114 + 1 ---CACCCACCCACUUCAGUGGCACUUGCCACCGCUUUGCAGCCAUUGUCGUUCCACUUUAGAAUUAUUUGCACAAAAUCUUUCGGCGGCAGCAAGGAUUUCUCGCUGCUGUGCUUC ---...............(((((....)))))......((((((...((......))....(((..((((.....))))..))))))((((((.((.....)).)))))).)))... ( -27.60) >consensus ___CACCCACCCACUUCAGUGGCACUUGCCACCGCUUUGCUGCCAUUGUCGUUCCACUUUAGAAUUAUUUGCACAAAAUCUUUCGGCGGCAGCAAGGAUUUCUCGCUACUGUGCUUC ...........(((....(((((....)))))..((((((((((...((((.....)....(((..((((.....))))..)))))))))))))))).............))).... (-28.30 = -28.55 + 0.25)

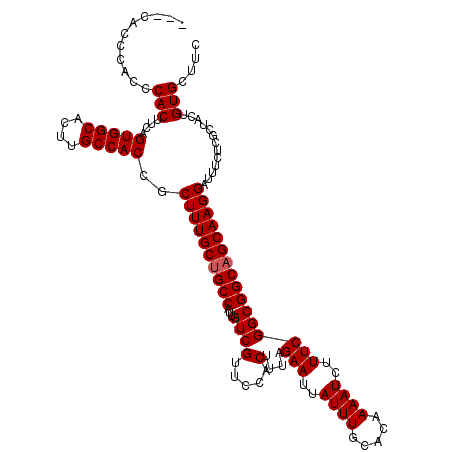
| Location | 20,345,415 – 20,345,530 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -35.51 |
| Energy contribution | -35.13 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20345415 115 - 22224390 GAAGCACAGUAGCGAGAAAUCCUUGCUGCCGCCGAAAGAUUUUGUGCAAAUAAUUCUAAAGUGGAACGACAAUGGCAGCAAAGCGGUGGCAAGUGCCACUGAAGUGGGUGGGUGG-- ....(((.((((((((.....))))))))((((......((((((((......((((.....))))........))).)))))(((((((....))))))).....)))).))).-- ( -37.04) >DroSec_CAF1 13210 117 - 1 GAAGCACAGUAGCGAGAAAUCCUUGCUGCCGCCGAAAGAUUUUGUGCAAAUAAUUCUAAAGUGAAACGACAAUGGCAGCAAAGCGGUGGCAAGUGCCACUGAAGUGGGUGGGUGGGU ...(((((((((((((.....))))))))...(....)....)))))......(((......)))....((.(.((..((...(((((((....)))))))...)).)).).))... ( -36.40) >DroSim_CAF1 14998 116 - 1 GAAGCACAGUAGCGAGAAAUCCUUGCUGCCGCCGAAAGAUUUUGUGCAAAUAAUUCUAAAGUGGAACGACAAUGGCAGCAAAGCGGUGGCAAGUGCCACUGAAGUGGGUGGGUG-GU ....(((.((((((((.....))))))))((((......((((((((......((((.....))))........))).)))))(((((((....))))))).....)))).)))-.. ( -37.04) >DroEre_CAF1 9932 114 - 1 GAAGCACAGCAGCGAGAAAUCCUUGCUGCCGCCGAAAGAUUUUGUGCAAAUAAUUCUAAAGUGGAACGACAAUGGCUGCAAAGCGGUGGCAAGUGCCACUGAAGUGGGUGGGUG--- ...(((((((((((((.....))))))))...(....)....)))))......((((.....))))...((.(.(((.((...(((((((....)))))))...))))).).))--- ( -37.80) >consensus GAAGCACAGUAGCGAGAAAUCCUUGCUGCCGCCGAAAGAUUUUGUGCAAAUAAUUCUAAAGUGGAACGACAAUGGCAGCAAAGCGGUGGCAAGUGCCACUGAAGUGGGUGGGUG___ ....(((.((((((((.....))))))))((((......((((((((......(((......))).........)).))))))(((((((....))))))).....)))).)))... (-35.51 = -35.13 + -0.37)



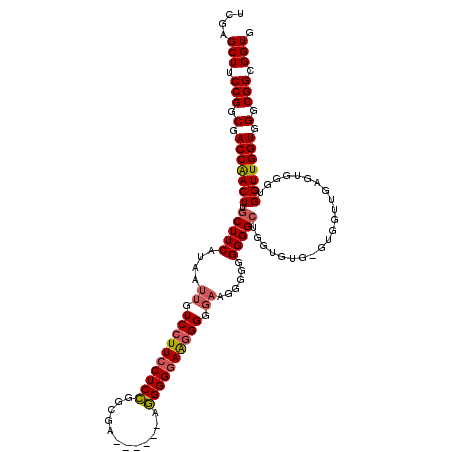
| Location | 20,345,530 – 20,345,629 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -44.20 |
| Consensus MFE | -33.92 |
| Energy contribution | -34.70 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20345530 99 + 22224390 UCGAGCUUCCGGCGACCGACUUGCUUCAUAAUUGUCCUUCCUCUGGCGU------AAGGGAAGGGGGGAAG---GGGGGAUGGUG------------UGGGUGGUUGGUGGGCGGCGGUG ....(((((((.(((((.(((..(.((((...(.((((((((((..(..------....)...))))))))---)).).)))).)------------..)))))))).)))).))).... ( -40.50) >DroSec_CAF1 13327 120 + 1 UCGAGCUUCCGGCGACCAACUUGCUUCAUAAUUGUCCUUCCUCCGGCGAUGGUGGAGGGGGAAGGGGGAAGGGGGGGGGCUGGUGUGUGUGGUUGAGUGGGUGGUUGGUGGGCGGCGGUG ....(((((((.((((((.((..(((((((.((.((((((((((.((....))...)))))))))).)).........((....)).))))...)))..)))))))).)))).))).... ( -44.50) >DroSim_CAF1 15114 109 + 1 UCGAGCUUCCGGCGACCAACUUGCUUCAUAAUUGUCCUUCCUCCGGCGA------AGGGGGAACGG-----GGGGGGGGCUGGUGUGGGUGGUUGAGUGGGUGGUUGGUGGGCGGCGGUG ....(((((((.((((((.((..(((((((...((((((((((((....------........)))-----)))))))))...)))).......)))..)))))))).)))).))).... ( -47.61) >consensus UCGAGCUUCCGGCGACCAACUUGCUUCAUAAUUGUCCUUCCUCCGGCGA______AGGGGGAAGGGGGAAGGGGGGGGGCUGGUGUG_GUGGUUGAGUGGGUGGUUGGUGGGCGGCGGUG ....(((.(((.(.(((((((.(((((....((.((((((((((............)))))))))).))......)))))......................))))))).).))).))). (-33.92 = -34.70 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:48 2006