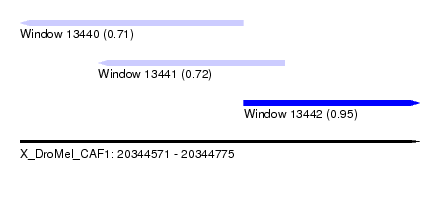
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,344,571 – 20,344,775 |
| Length | 204 |
| Max. P | 0.952874 |

| Location | 20,344,571 – 20,344,685 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -25.82 |
| Energy contribution | -26.66 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20344571 114 - 22224390 ---G---GUUGUAAGGACAUGCGAGGACAUCCGCUGGGAUAAUAAUAAACAUCGGGGGACUAAAGUUGUGGACAGGCGGAGGGAGGGGGAACACCCCCAAGUCAAGUAAUUUGUCAACGA ---.---.((((...(((((((...(((.((((((.(..((......(((.(..(....)..).))).))..).))))))....(((((....)))))..)))..)))...)))).)))) ( -34.90) >DroSec_CAF1 12317 116 - 1 CGGG---GUUGUAAGGACAUGCGAGGACAUCCGCUGGGAUAAUAAUAAACAUCGGGGGACUGAAGUGGUGGACAGGAGGAGGGAGGGGG-ACACCCCCAAGUCAAGUAAUUUGUCAACGA ....---.((((...(((((((...(((.((((((.............((.((((....)))).)))))))).....((.(((..(...-.).)))))..)))..)))...)))).)))) ( -32.71) >DroSim_CAF1 14109 116 - 1 CGGG---GUUGUAAGGACAUGCGAGGACAUCCGCUGGGAUAAUAAUAAACAUCGGGGGACUGAAGUUGUGGACAGGAGGAGGGAGGGGG-ACACCCCCAAGUCAAGUAAUUUGUCAACGA ....---.((((...(((((((...(((.(((.(((...((......(((.((((....)))).))).))..)))..)))....(((((-...)))))..)))..)))...)))).)))) ( -33.20) >DroEre_CAF1 8971 111 - 1 AGGGGGAGGCUGGAGGACAUGCGAGGACAUCCGCUGGGAUAAUAAUAAACAUCAGGGGACUGAAGUCGUG-ACA-----AGGGA-GGGG-AGACCCCC-AGUCAAGUAAUUUGUCAACGA ...............(((((((...(((((((....))))...........((((....)))).))).((-((.-----..((.-(((.-...)))))-.)))).)))...))))..... ( -33.70) >DroYak_CAF1 12957 109 - 1 CGGG---GGCUGGGGGACAUGCGAGGACAUCCGCAGGGAUAAUAAUAAACAUCGGGGGACUGAAGUUGUGGACA-----AGGAA-GGGG-ACACCCCC-AGUCAAGUAAUUUGUCAACGA ....---(((((((((.(.(((.(.(((((((....))))...........((((....)))).))).).).))-----.)...-(...-.).)))))-))))................. ( -36.00) >consensus CGGG___GUUGUAAGGACAUGCGAGGACAUCCGCUGGGAUAAUAAUAAACAUCGGGGGACUGAAGUUGUGGACAGG_GGAGGGAGGGGG_ACACCCCCAAGUCAAGUAAUUUGUCAACGA ...............(((((((...(((((((....)))).......(((.((((....)))).))).................(((((....)))))..)))..)))...))))..... (-25.82 = -26.66 + 0.84)


| Location | 20,344,611 – 20,344,706 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 85.53 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -23.70 |
| Energy contribution | -25.07 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20344611 95 - 22224390 -GGGACGCCCUCCGGAGGG-AGG----------GGUUGUAAGGACAUGCGAGGACAUCCGCUGGGAUAAUAAUAAACAUCGGGGGACUAAAGUUGUGGACAGGCGGA -.(..(.((((((....))-)))----------).(((((......))))))..).((((((.(..((......(((.(..(....)..).))).))..).)))))) ( -28.50) >DroSec_CAF1 12356 105 - 1 -GGGACUCCCUCCGUUGGG-AGGGGGUGUGCGGGGUUGUAAGGACAUGCGAGGACAUCCGCUGGGAUAAUAAUAAACAUCGGGGGACUGAAGUGGUGGACAGGAGGA -.(.(((((((((....))-))))))).)(((..(((.....))).)))..(..((.(((((.(..((....))..).((((....)))))))))))..)....... ( -34.80) >DroSim_CAF1 14148 105 - 1 -GGGACUCCCUCCGUUGGG-AGGGGGUGUGCGGGGUUGUAAGGACAUGCGAGGACAUCCGCUGGGAUAAUAAUAAACAUCGGGGGACUGAAGUUGUGGACAGGAGGA -.(.(((((((((....))-))))))).)(((((.(((((......))))).....))))).............(((.((((....)))).)))............. ( -34.00) >DroYak_CAF1 12994 102 - 1 AGGGACCCCCUCCGUAGGGAAGGGGGUGUGCGGGGGCUGGGGGACAUGCGAGGACAUCCGCAGGGAUAAUAAUAAACAUCGGGGGACUGAAGUUGUGGACA-----A ....(((((((((....)).))))))).((((((..((.(....)......))...))))))............(((.((((....)))).))).......-----. ( -35.30) >consensus _GGGACUCCCUCCGUAGGG_AGGGGGUGUGCGGGGUUGUAAGGACAUGCGAGGACAUCCGCUGGGAUAAUAAUAAACAUCGGGGGACUGAAGUUGUGGACAGGAGGA ....(((((((((....)).)))))))..............(..(((........((((....)))).......(((.((((....)))).))))))..)....... (-23.70 = -25.07 + 1.37)

| Location | 20,344,685 – 20,344,775 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -21.36 |
| Consensus MFE | -13.07 |
| Energy contribution | -13.25 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20344685 90 + 22224390 -------CCU-CCCUCCGGAGGGCGUCCC-CCUCGGACUCAGGCUAAUAAAUAAUAAACAAACCUUGUCCCAUA--GACAGGCUUCCACUGCCUCCACCCC -------(((-...((((..(((....))-)..))))...)))......................((((.....--))))(((.......)))........ ( -20.50) >DroSec_CAF1 12433 99 + 1 CACACCCCCU-CCCAACGGAGGGAGUCCC-CCUCGGACUCAGGCUAAUAAAUAAUAAACAAACCUUGUCCCAUACCGACUGGCUUCCUCUACCUACCCCCC ....((((((-((....))))))((((..-....((((..(((...................))).))))......))))))................... ( -20.81) >DroSim_CAF1 14225 99 + 1 CACACCCCCU-CCCAACGGAGGGAGUCCC-CCUCGGACUCAGGCUAAUAAAUAAUAAACAAACCUUGUCCCAUACCGACUGGCUUCCUCUACCUACCCCCC ....((((((-((....))))))((((..-....((((..(((...................))).))))......))))))................... ( -20.81) >DroYak_CAF1 13066 88 + 1 CACACCCCCUUCCCUACGGAGGGGGUCCCUCCUCGGGCUGAGGCUAAUAAAUAAUAAACAAACCUUGUCCCAUGCCGACUGA-------------CUCCCC ...(((((((((.....)))))))))........(((.(((((...................))))).)))...........-------------...... ( -23.31) >consensus CACACCCCCU_CCCAACGGAGGGAGUCCC_CCUCGGACUCAGGCUAAUAAAUAAUAAACAAACCUUGUCCCAUACCGACUGGCUUCCUCUACCUACCCCCC ......((((.((....))))))((((.......((((..(((...................))).))))......))))..................... (-13.07 = -13.25 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:45 2006