
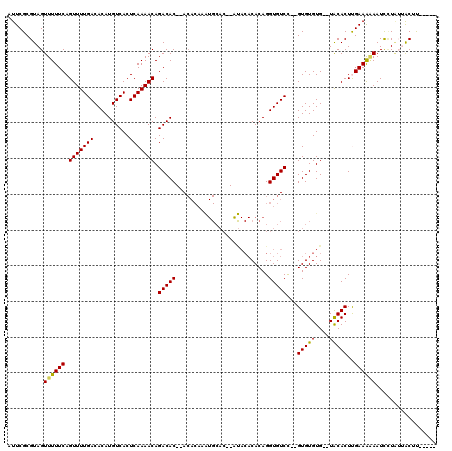
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,344,296 – 20,344,443 |
| Length | 147 |
| Max. P | 0.998519 |

| Location | 20,344,296 – 20,344,405 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -14.23 |
| Energy contribution | -13.87 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20344296 109 + 22224390 AUUCGCGUAGUUUUUCAGUUUUGACACAUGUCACUCAAAACAGACAC--ACACAAAUGCAC--GUACACACAGGUGUCCGUGUGUGUG--UACACUCGAAAGAAUCUUAUUACUU----- ......(((((..(((.(((((((..........)))))))..((((--(((((...((((--.(......).))))...))))))))--)..........)))....)))))..----- ( -27.80) >DroSec_CAF1 12040 91 + 1 AUUCGCGUAGUUUUUCAGUUUUGACACAUGUCACUCAAAACAGACAC--A------------------CACAGGUGUCU--GUGUGUG--UACACUCGAAGAAAUCGUAUUACUU----- ....(((...((((((.(((((((..........)))))))..((((--(------------------(((((....))--)))))))--)......))))))..))).......----- ( -28.20) >DroSim_CAF1 13813 107 + 1 AUUCGCGUAGUUUUUCAGUUUUGACACAUGUCACUCAAAACAGACAC--ACACAAAUGCAC--GUACACACAGGUGUCU--GUGUGUG--UACACUUGAAGAAAUCCUAUUACUU----- ......((((..((((.....((((....)))).((((.....((((--(((((...((((--.(......).)))).)--)))))))--)....)))).))))..)))).....----- ( -27.90) >DroEre_CAF1 8706 113 + 1 AUUCGCGUAGUUUUUCAGUUUUGACACAUGUCACUCAAAACAGACACGCACACAAGUGCACGCACACACACAGGUGUCC--GUGUGUG--UGCACUUGAAAGAAUUUUCAGGCUUAU--- ((((((((.((((....(((((((..........)))))))))))))))...((((((((((((((((((....)))..--)))))))--))))))))...))))............--- ( -40.80) >DroYak_CAF1 12684 114 + 1 AUUCGCGUAGUUUUUCAGUUUUGACACAUGUCACUCAAAACAGACAC--ACACAAAUGCAC--AUACACACAGGUGUCC--GUGUGUGCGUACACUUGAAAUAAUCCUGAGGCGCAUAAC ....((((.....(((((..(((.....((((..........)))).--......((((((--(((((((....)))..--))))))))))..........)))..)))))))))..... ( -27.20) >consensus AUUCGCGUAGUUUUUCAGUUUUGACACAUGUCACUCAAAACAGACAC__ACACAAAUGCAC__AUACACACAGGUGUCC__GUGUGUG__UACACUUGAAAAAAUCCUAUUACUU_____ ..........((((((.(((((((..........))))))).(((((..........................)))))...(((((....)))))..))))))................. (-14.23 = -13.87 + -0.36)


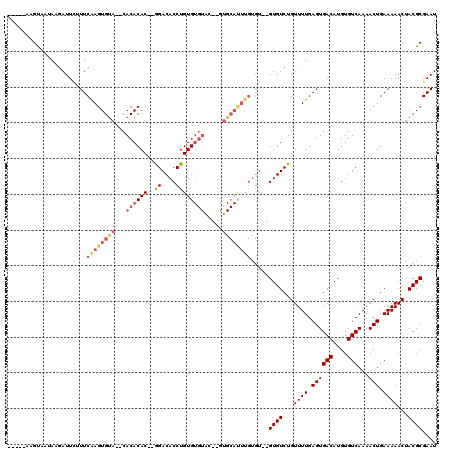
| Location | 20,344,296 – 20,344,405 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -33.84 |
| Consensus MFE | -18.04 |
| Energy contribution | -19.64 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20344296 109 - 22224390 -----AAGUAAUAAGAUUCUUUCGAGUGUA--CACACACACGGACACCUGUGUGUAC--GUGCAUUUGUGU--GUGUCUGUUUUGAGUGACAUGUGUCAAAACUGAAAAACUACGCGAAU -----..........((((...((((((((--(..((((((((....))))))))..--)))))))))...--((((...((((.((((((....)))...))).))))...)))))))) ( -32.20) >DroSec_CAF1 12040 91 - 1 -----AAGUAAUACGAUUUCUUCGAGUGUA--CACACAC--AGACACCUGUG------------------U--GUGUCUGUUUUGAGUGACAUGUGUCAAAACUGAAAAACUACGCGAAU -----........(((.....))).(((((--(((((((--((....)))))------------------)--)))((.(((((((..........))))))).)).....))))).... ( -26.40) >DroSim_CAF1 13813 107 - 1 -----AAGUAAUAGGAUUUCUUCAAGUGUA--CACACAC--AGACACCUGUGUGUAC--GUGCAUUUGUGU--GUGUCUGUUUUGAGUGACAUGUGUCAAAACUGAAAAACUACGCGAAU -----............(((..((((((((--(((((((--((....))))))))..--)))))))))...--((((...((((.((((((....)))...))).))))...))))))). ( -31.60) >DroEre_CAF1 8706 113 - 1 ---AUAAGCCUGAAAAUUCUUUCAAGUGCA--CACACAC--GGACACCUGUGUGUGUGCGUGCACUUGUGUGCGUGUCUGUUUUGAGUGACAUGUGUCAAAACUGAAAAACUACGCGAAU ---....((.(((((....))))).))(((--(((((((--((....))))))))))))..((((....))))((((...((((.((((((....)))...))).))))...)))).... ( -40.90) >DroYak_CAF1 12684 114 - 1 GUUAUGCGCCUCAGGAUUAUUUCAAGUGUACGCACACAC--GGACACCUGUGUGUAU--GUGCAUUUGUGU--GUGUCUGUUUUGAGUGACAUGUGUCAAAACUGAAAAACUACGCGAAU ....((((.(((((((((((..((((((((((.((((((--((....)))))))).)--)))))))))...--)))...))))))))((((....))))..............))))... ( -38.10) >consensus _____AAGUAAUAAGAUUCUUUCAAGUGUA__CACACAC__GGACACCUGUGUGUAC__GUGCAUUUGUGU__GUGUCUGUUUUGAGUGACAUGUGUCAAAACUGAAAAACUACGCGAAU ......................((((((((...((((((..((....)))))))).....)))))))).....((((...((((.((((((....)))...))).))))...)))).... (-18.04 = -19.64 + 1.60)


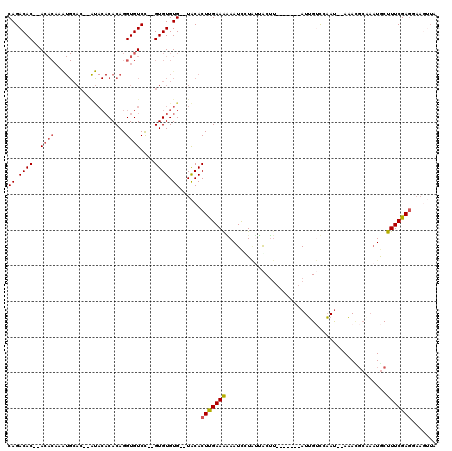
| Location | 20,344,336 – 20,344,443 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.20 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -7.67 |
| Energy contribution | -7.99 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.27 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20344336 107 + 22224390 CAGACAC--ACACAAAUGCAC--GUACACACAGGUGUCCGUGUGUGUG--UACACUCGAAAGAAUCUUAUUACUU-------AUGGUCCCAUAUAUAUGCGAAUGCUUUCGAGGAAGUUA ..(((((--((((....((((--(.((((....)))).))))))))))--)...((((((((..((.(((....(-------(((....))))...))).))...))))))))...))). ( -34.50) >DroSec_CAF1 12080 87 + 1 CAGACAC--A------------------CACAGGUGUCU--GUGUGUG--UACACUCGAAGAAAUCGUAUUACUU-------AUUGUUCAAU--AAACGCAAAUGCUUUCGAGGAAGUUA ...((((--(------------------(((((....))--)))))))--)...(((((((((...(((.....)-------))..)))...--....((....)).))))))....... ( -25.30) >DroSim_CAF1 13853 103 + 1 CAGACAC--ACACAAAUGCAC--GUACACACAGGUGUCU--GUGUGUG--UACACUUGAAGAAAUCCUAUUACUU-------AUUGUUCAAU--AAACGCAAAUGCUUUCGAGUAAGUUA ...((((--(((((...((((--.(......).)))).)--)))))))--)..((((((((............((-------(((....)))--))..((....))))))))))...... ( -24.90) >DroEre_CAF1 8746 101 + 1 CAGACACGCACACAAGUGCACGCACACACACAGGUGUCC--GUGUGUG--UGCACUUGAAAGAAUUUUCAGGCUUAU-----AUUGGCUAAU--AAAUGCGAAUACUUUCGA-------- ......((((..((((((((((((((((((....)))..--)))))))--))))))))............((((...-----...))))...--...))))...........-------- ( -36.80) >DroYak_CAF1 12724 106 + 1 CAGACAC--ACACAAAUGCAC--AUACACACAGGUGUCC--GUGUGUGCGUACACUUGAAAUAAUCCUGAGGCGCAUAACCUAUUGACAAAC--AAA------UACUUUCGAGUAAUAUG .......--......((((((--(((((((....)))..--))))))))))..((((((((........(((.......))).(((.....)--)).------...))))))))...... ( -22.20) >consensus CAGACAC__ACACAAAUGCAC__AUACACACAGGUGUCC__GUGUGUG__UACACUUGAAAAAAUCCUAUUACUU_______AUUGUCCAAU__AAACGCAAAUGCUUUCGAGGAAGUUA ((.((((..((((....................))))....)))).))......(((((((.............................................)))))))....... ( -7.67 = -7.99 + 0.32)


| Location | 20,344,336 – 20,344,443 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.20 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -10.02 |
| Energy contribution | -9.94 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.33 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20344336 107 - 22224390 UAACUUCCUCGAAAGCAUUCGCAUAUAUAUGGGACCAU-------AAGUAAUAAGAUUCUUUCGAGUGUA--CACACACACGGACACCUGUGUGUAC--GUGCAUUUGUGU--GUGUCUG .......((((((((...((...(((.((((....)))-------).)))....))..))))))))..((--((((((((((.((((....)))).)--)))....)))))--))).... ( -32.10) >DroSec_CAF1 12080 87 - 1 UAACUUCCUCGAAAGCAUUUGCGUUU--AUUGAACAAU-------AAGUAAUACGAUUUCUUCGAGUGUA--CACACAC--AGACACCUGUG------------------U--GUGUCUG .......((((((.((....))((((--(((.......-------.))))).))......))))))...(--(((((((--((....)))))------------------)--))))... ( -24.70) >DroSim_CAF1 13853 103 - 1 UAACUUACUCGAAAGCAUUUGCGUUU--AUUGAACAAU-------AAGUAAUAGGAUUUCUUCAAGUGUA--CACACAC--AGACACCUGUGUGUAC--GUGCAUUUGUGU--GUGUCUG ..(((((.((((((((......))))--.))))....)-------))))....((((.....((((((((--(((((((--((....))))))))..--)))))))))...--..)))). ( -29.60) >DroEre_CAF1 8746 101 - 1 --------UCGAAAGUAUUCGCAUUU--AUUAGCCAAU-----AUAAGCCUGAAAAUUCUUUCAAGUGCA--CACACAC--GGACACCUGUGUGUGUGCGUGCACUUGUGUGCGUGUCUG --------..(((((..((((..(((--((........-----)))))..))))....)))))....(((--(((((((--((....))))))))))))..((((....))))....... ( -34.40) >DroYak_CAF1 12724 106 - 1 CAUAUUACUCGAAAGUA------UUU--GUUUGUCAAUAGGUUAUGCGCCUCAGGAUUAUUUCAAGUGUACGCACACAC--GGACACCUGUGUGUAU--GUGCAUUUGUGU--GUGUCUG ((...((((....))))------..)--).........((((.....))))..((((.....((((((((((.((((((--((....)))))))).)--)))))))))...--..)))). ( -30.10) >consensus UAACUUACUCGAAAGCAUUCGCAUUU__AUUGGACAAU_______AAGUAAUAAGAUUCUUUCAAGUGUA__CACACAC__GGACACCUGUGUGUAC__GUGCAUUUGUGU__GUGUCUG .........(....)..................................................(((......)))....((((((....(((((....)))))........)))))). (-10.02 = -9.94 + -0.08)

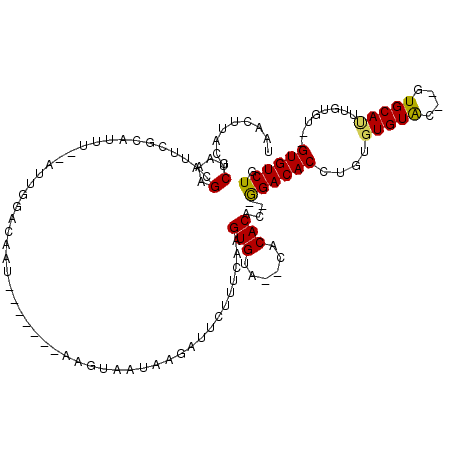

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:42 2006