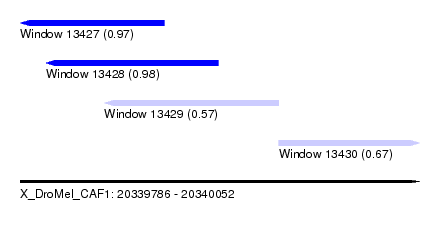
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,339,786 – 20,340,052 |
| Length | 266 |
| Max. P | 0.978975 |

| Location | 20,339,786 – 20,339,882 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -18.66 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20339786 96 - 22224390 UGCGUUUGAAUUCUCCAGCAAGCUUUUACUUUAAUUGCCCCAAAAGAACUUGGGAUAUACAAAUAAUAUGGAUCCUAAG-------------AU-------U---U-CGCAAUCGAACAG ...((((((........((((.............)))).......(((((((((((...((.......)).))))))))-------------..-------)---)-)....)))))).. ( -18.02) >DroSec_CAF1 7680 103 - 1 UGCGUUUGAAUUCUCCAUCAAGCUUUUACUUUAAUUGCCCCAAAAGAACUUGGGGUAUACAAAGUAUAUGGGUCCUGAG-------------AACCAUCUAU---U-CGCAAUCAAAGAG ((((...((.(((((......((((.((((((...((((((((......))))))))...))))))...))))...)))-------------))...))...---.-))))......... ( -27.00) >DroSim_CAF1 9209 106 - 1 UGCGUUUGAAUUCUCCAUCAAGCUUUUACUUUAAUUGCCCCAAAAGAACUUGGGGUAUACAAAGUAUAUGGAUCCUAAG-------------AACCAUCUAUUUCU-CGCAAUCAAACAG (((((((((........))))))...((((((...((((((((......))))))))...)))))).((((.((....)-------------).))))........-.)))......... ( -27.00) >DroEre_CAF1 4003 119 - 1 UGCGUUUGAAUUCUUCAGCAAGCUUUUACUUUAAUUGCCCCAAAACAACUUGGGGUAUACAAAGUGUACGGGCCCUAAGAUCUCGGCGUGAGAACUAACUAUUUUU-UGCAAUCGAGCAG ((((((.((..((((.((...((((.((((((...((((((((......))))))))...))))))...)))).))))))..)))))(..((((........))))-..)......))). ( -30.70) >DroYak_CAF1 7985 118 - 1 UGCGUUUGAAUUCUCCAGCAAGCUUUUACUUUAAUUGCCCCAACAGAACUCGGGGUAUACAAAGUAUUUGGCUCCUAAGAUAUCGGAGAGAGAACUAUC--UUUUUCUGCAAUCGAACGG ..(((((((..(((..((..((((..((((((...((((((..........))))))...))))))...)))).)).)))...((((((((((....))--))))))))...))))))). ( -36.60) >consensus UGCGUUUGAAUUCUCCAGCAAGCUUUUACUUUAAUUGCCCCAAAAGAACUUGGGGUAUACAAAGUAUAUGGAUCCUAAG_____________AACCAUCUAUUU_U_CGCAAUCGAACAG ((((((((..........)))))...((((((...((((((((......))))))))...))))))..........................................)))......... (-18.66 = -19.30 + 0.64)


| Location | 20,339,803 – 20,339,918 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -21.88 |
| Energy contribution | -22.36 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.978975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20339803 115 - 22224390 GACUUACAGCCCCUCAC----CCACACACUCGCAUCUCGAUGCGUUUGAAUUCUCCAGCAAGCUUUUACUUUAAUUGCCCCAAAAGAACUUGGGAUAUACAAAUAAUAUGGAUCCUAAG- ..((((...........----.....((..(((((....)))))..)).....((((((((.............))))(((((......)))))..............))))...))))- ( -19.62) >DroSec_CAF1 7704 113 - 1 GACUUACAGC--CUCAC----CCACACACUCGCAUCUCGAUGCGUUUGAAUUCUCCAUCAAGCUUUUACUUUAAUUGCCCCAAAAGAACUUGGGGUAUACAAAGUAUAUGGGUCCUGAG- ......(((.--...((----(((.......((((....))))((((((........))))))...((((((...((((((((......))))))))...))))))..))))).)))..- ( -33.40) >DroSim_CAF1 9236 113 - 1 GACUUACAGC--CUCAC----ACACACACUCGCAUCUCGAUGCGUUUGAAUUCUCCAUCAAGCUUUUACUUUAAUUGCCCCAAAAGAACUUGGGGUAUACAAAGUAUAUGGAUCCUAAG- ..((((...(--(....----..........((((....))))((((((........))))))...((((((...((((((((......))))))))...))))))...))....))))- ( -27.40) >DroEre_CAF1 4042 114 - 1 GACUUACUGC--CUCAC----CCACACACUCGCAUCUCGAUGCGUUUGAAUUCUUCAGCAAGCUUUUACUUUAAUUGCCCCAAAACAACUUGGGGUAUACAAAGUGUACGGGCCCUAAGA ..((((..((--((...----.....((..(((((....)))))..))..................((((((...((((((((......))))))))...))))))...))))..)))). ( -31.90) >DroYak_CAF1 8023 118 - 1 GACUUACAGC--AUCACCCACCCACACACUCGCAUCUCGAUGCGUUUGAAUUCUCCAGCAAGCUUUUACUUUAAUUGCCCCAACAGAACUCGGGGUAUACAAAGUAUUUGGCUCCUAAGA ..((((....--..............((..(((((....)))))..))............((((..((((((...((((((..........))))))...))))))...))))..)))). ( -27.20) >consensus GACUUACAGC__CUCAC____CCACACACUCGCAUCUCGAUGCGUUUGAAUUCUCCAGCAAGCUUUUACUUUAAUUGCCCCAAAAGAACUUGGGGUAUACAAAGUAUAUGGAUCCUAAG_ ..((((....................((..(((((....)))))..))..............((..((((((...((((((((......))))))))...))))))...))....)))). (-21.88 = -22.36 + 0.48)

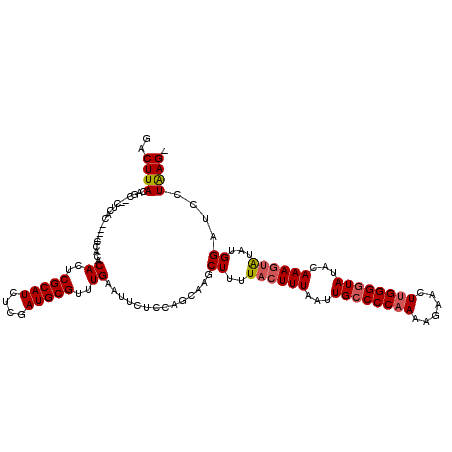
| Location | 20,339,842 – 20,339,958 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.36 |
| Mean single sequence MFE | -19.97 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.32 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20339842 116 - 22224390 UCCUGCGCUGCCCUCUGUCUGCGUGUAAUUAAAUAAAAACGACUUACAGCCCCUCAC----CCACACACUCGCAUCUCGAUGCGUUUGAAUUCUCCAGCAAGCUUUUACUUUAAUUGCCC ....((((.((.....))..))))((((((((((((((...................----..........((((....))))(((((..........)))))))))).))))))))).. ( -18.30) >DroSec_CAF1 7743 114 - 1 UCCUGCGCUGUCCUCUGUCUGCGUGUAAUUAAGUAAAAACGACUUACAGC--CUCAC----CCACACACUCGCAUCUCGAUGCGUUUGAAUUCUCCAUCAAGCUUUUACUUUAAUUGCCC ....((((.(........).))))((((((((((((((............--.....----..........((((....))))((((((........)))))))))))).)))))))).. ( -22.80) >DroSim_CAF1 9275 114 - 1 UCCUGCGCUGCCCUCUGUCUGCGUGUAAUUAAAUAAAAACGACUUACAGC--CUCAC----ACACACACUCGCAUCUCGAUGCGUUUGAAUUCUCCAUCAAGCUUUUACUUUAAUUGCCC ....((((.((.....))..))))((((((((((((((............--.....----..........((((....))))((((((........))))))))))).))))))))).. ( -20.60) >DroEre_CAF1 4082 114 - 1 UCCUGCGCUGCCCUCUGUCUGCGUGUAAUUAAAUAAAAACGACUUACUGC--CUCAC----CCACACACUCGCAUCUCGAUGCGUUUGAAUUCUUCAGCAAGCUUUUACUUUAAUUGCCC ....((((.((.....))..))))((((((((((((((..(.(((.(((.--.....----.....((..(((((....)))))..)).......))).))))))))).))))))))).. ( -19.07) >DroYak_CAF1 8063 118 - 1 UCCUGCGCUGCCCUCUGUCUGCGUGUAAUUAAAUAAAAACGACUUACAGC--AUCACCCACCCACACACUCGCAUCUCGAUGCGUUUGAAUUCUCCAGCAAGCUUUUACUUUAAUUGCCC ....((((.((.....))..))))((((((((((((((..........((--(((.......................)))))(((((..........)))))))))).))))))))).. ( -19.10) >consensus UCCUGCGCUGCCCUCUGUCUGCGUGUAAUUAAAUAAAAACGACUUACAGC__CUCAC____CCACACACUCGCAUCUCGAUGCGUUUGAAUUCUCCAGCAAGCUUUUACUUUAAUUGCCC ....((((............))))((((((((((((((.................................((((....))))(((((..........)))))))))).))))))))).. (-18.48 = -18.32 + -0.16)



| Location | 20,339,958 – 20,340,052 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 98.58 |
| Mean single sequence MFE | -12.27 |
| Consensus MFE | -11.63 |
| Energy contribution | -11.63 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20339958 94 + 22224390 AAAAGGACAUUUGAAGCGUUGUACAUAAAUGAAAAUGAAAUAAAUUAAAAUAAGUGUCGAAGCUGAAGGCAAAGGCAAAGGCAAAAAGGACCAA .....((((((((...((((.......))))..(((.......)))....))))))))...(((....((....))...)))............ ( -12.20) >DroSec_CAF1 7857 94 + 1 AAAAGGACAUUUGAAGCGUUGUGCAUAAAUGAAAAUGAAAUAAAUUAAAAUAAGUGUCGAAGCUAAAGGCAAAGGCAAAGGCAAAAAGGACCAA .....((((((((..((.....))....((....))..............))))))))...(((....((....))...)))............ ( -12.30) >DroSim_CAF1 9389 94 + 1 AAAAGGACAUUUGAAGCGUUGUGCAUAAAUGAAAAUGAAAUAAAUUAAAAUAAGUGUCGAAGCUAAAGGCAAAGGCAAAGGCAAAAAGGACCAA .....((((((((..((.....))....((....))..............))))))))...(((....((....))...)))............ ( -12.30) >consensus AAAAGGACAUUUGAAGCGUUGUGCAUAAAUGAAAAUGAAAUAAAUUAAAAUAAGUGUCGAAGCUAAAGGCAAAGGCAAAGGCAAAAAGGACCAA .....((((((((.....((((.(((........)))..)))).......))))))))...(((....((....))...)))............ (-11.63 = -11.63 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:34 2006