
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,273,467 – 20,273,627 |
| Length | 160 |
| Max. P | 0.999918 |

| Location | 20,273,467 – 20,273,587 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -31.82 |
| Energy contribution | -32.22 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20273467 120 + 22224390 ACAUCUCUAGCUGCCACCCACUCCAUCCUGGACUUUGUGCGUAGCACUCCGCGGAGGGCGCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUC .........((((((((....(((.....)))....))).))))).......(((((((((((((....)))))).........((.(((((...))))))).......))).))))... ( -45.00) >DroPse_CAF1 12248 110 + 1 ----------CUGCCACCCACUCGAUACUGGACUUUGUGCGGAGCACACCGCGUCGCGCCCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCGAACAAGAGCCUCUCCAUC ----------..........(((....((((.....(((((..((.....))..))))))))).((((((((((((((......)).))))))...)))))).....))).......... ( -36.60) >DroSec_CAF1 3191 120 + 1 ACAUCUCCAGCUGCCACCCACUCCAUCCUGGACUUUGUGCGUAGCACUCCGCGGAGGGCGCAGUUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUC .........((((((((....(((.....)))....))).))))).......(((((((...((((((((((((((((......)).))))))...)))).))))....))).))))... ( -46.80) >DroSim_CAF1 3494 120 + 1 ACAUCUCCAGCUGCCACUCACUCCAUCCUGGACUUUGUGCGUAGCACUCCGCGGAGGGCGCAGUUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUC .........((((((((....(((.....)))....))).))))).......(((((((...((((((((((((((((......)).))))))...)))).))))....))).))))... ( -47.50) >DroPer_CAF1 12087 110 + 1 ----------CUGCCACCCACUCGAUACUGGACUUUGUGCGGAGCACACCGCGUCGCGCCCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCGAACAAGAGCCUCUCCAUC ----------..........(((....((((.....(((((..((.....))..))))))))).((((((((((((((......)).))))))...)))))).....))).......... ( -36.60) >consensus ACAUCUC_AGCUGCCACCCACUCCAUCCUGGACUUUGUGCGUAGCACUCCGCGGAGGGCGCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUC .........((((((.......((.(((((((....((((...)))))))).))).)).((((((....)))))).........))))))...((..((((......)))).))...... (-31.82 = -32.22 + 0.40)

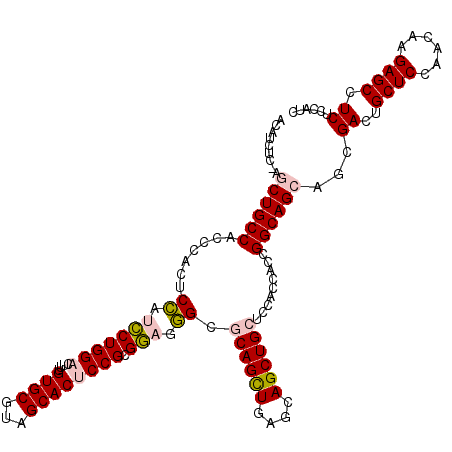

| Location | 20,273,467 – 20,273,587 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -51.42 |
| Consensus MFE | -39.38 |
| Energy contribution | -41.14 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20273467 120 - 22224390 GAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUCAGCUGCGCCCUCCGCGGAGUGCUACGCACAAAGUCCAGGAUGGAGUGGGUGGCAGCUAGAGAUGU ......((.(((((....)))))..))(((((..((.((((.((.((((((....))))))..)).))))...((((...))))..(.(((.....))).).))..)))))......... ( -52.10) >DroPse_CAF1 12248 110 - 1 GAUGGAGAGGCUCUUGUUCGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUCAGCUGGGCGCGACGCGGUGUGCUCCGCACAAAGUCCAGUAUCGAGUGGGUGGCAG---------- .........(((((..((((((((((.(((.(..((...))..)))).))))))).(((((((.....((((......)))).....)))))))...)))..)).)))..---------- ( -49.80) >DroSec_CAF1 3191 120 - 1 GAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUCAACUGCGCCCUCCGCGGAGUGCUACGCACAAAGUCCAGGAUGGAGUGGGUGGCAGCUGGAGAUGU ...((((.(((.(..((((.((((((.(((.(..((...))..)))).)))))))))).).))))))).(((..((((((.(((....(((.....)))))).)))))).)))....... ( -53.50) >DroSim_CAF1 3494 120 - 1 GAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUCAACUGCGCCCUCCGCGGAGUGCUACGCACAAAGUCCAGGAUGGAGUGAGUGGCAGCUGGAGAUGU ...((((.(((.(..((((.((((((.(((.(..((...))..)))).)))))))))).).))))))).(((..((((((.(((....(((.....)))))).)))))).)))....... ( -51.90) >DroPer_CAF1 12087 110 - 1 GAUGGAGAGGCUCUUGUUCGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUCAGCUGGGCGCGACGCGGUGUGCUCCGCACAAAGUCCAGUAUCGAGUGGGUGGCAG---------- .........(((((..((((((((((.(((.(..((...))..)))).))))))).(((((((.....((((......)))).....)))))))...)))..)).)))..---------- ( -49.80) >consensus GAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUCAGCUGCGCCCUCCGCGGAGUGCUACGCACAAAGUCCAGGAUGGAGUGGGUGGCAGCU_GAGAUGU (.(((((.(((.(..(((.(((((((.(((.(..((...))..)))).)))))))))).).)))))))))....((((((.(((....(((.....)))))).))))))........... (-39.38 = -41.14 + 1.76)


| Location | 20,273,507 – 20,273,627 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -52.94 |
| Consensus MFE | -48.18 |
| Energy contribution | -48.30 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.55 |
| SVM RNA-class probability | 0.999918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20273507 120 + 22224390 GUAGCACUCCGCGGAGGGCGCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUCGACCAGUUCCCGAUGGAGUCGCCGCUGAUCCUCCAGCAGC ..........((((((((..((((.(((((((((((((......)).))))))...)))))......(.((..((((((((.........))))))))..))))))).)))))).))... ( -52.70) >DroPse_CAF1 12278 120 + 1 GGAGCACACCGCGUCGCGCCCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCGAACAAGAGCCUCUCCAUCGAUCAGUAUCCGGUGGAGUCGCCGCUGCUGCUGCUGCAAC (((((.....(((...)))..((((....)))))))))..((.(((((((((((...((((......))))..((((((((.........))))))))....))))))))))).)).... ( -54.40) >DroSec_CAF1 3231 120 + 1 GUAGCACUCCGCGGAGGGCGCAGUUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUCGACCAGUUCCCGCUGGAGUCGCCGCUGCUCCUCCAGCAAC ..........((((((((.((((((....)))))).))......((((((.((((((((((......))))...........(((((....))))))))))).)))))).)))).))... ( -51.60) >DroSim_CAF1 3534 120 + 1 GUAGCACUCCGCGGAGGGCGCAGUUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUCGACCAGUUCCCGCUGGAGUCGCCGCUGCUCCUCCAGCAAC ..........((((((((.((((((....)))))).))......((((((.((((((((((......))))...........(((((....))))))))))).)))))).)))).))... ( -51.60) >DroPer_CAF1 12117 120 + 1 GGAGCACACCGCGUCGCGCCCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCGAACAAGAGCCUCUCCAUCGAUCAGUAUCCGGUGGAGUCGCCGCUGCUGCUGCUGCAAC (((((.....(((...)))..((((....)))))))))..((.(((((((((((...((((......))))..((((((((.........))))))))....))))))))))).)).... ( -54.40) >consensus GUAGCACUCCGCGGAGGGCGCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUCGACCAGUUCCCGCUGGAGUCGCCGCUGCUCCUCCAGCAAC ..........(((((((..((((((....)))))).........((((((.(((((.((((......))))...(((((((.........)))))))))))).))))))))))).))... (-48.18 = -48.30 + 0.12)

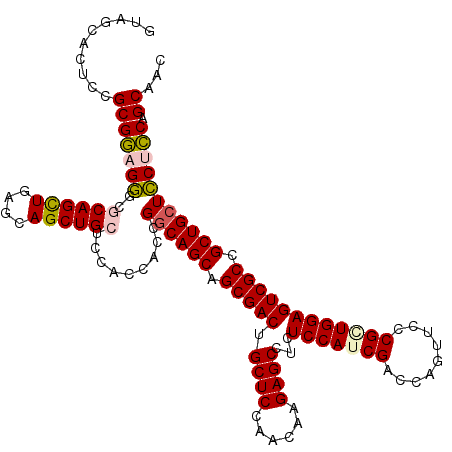
| Location | 20,273,507 – 20,273,627 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -60.64 |
| Consensus MFE | -52.58 |
| Energy contribution | -52.70 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20273507 120 - 22224390 GCUGCUGGAGGAUCAGCGGCGACUCCAUCGGGAACUGGUCGAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUCAGCUGCGCCCUCCGCGGAGUGCUAC .((((.(((((..((((((((((((((((((.......))))))))...(((((....))))).))))))))))...(..((.((((....)))).))..)..)))))))))........ ( -62.00) >DroPse_CAF1 12278 120 - 1 GUUGCAGCAGCAGCAGCGGCGACUCCACCGGAUACUGAUCGAUGGAGAGGCUCUUGUUCGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUCAGCUGGGCGCGACGCGGUGUGCUCC ....((.(.(((((((((((..(((((.(((.......))).)))))..((((......)))).))))))))))).).))..(((((((((((((.((.....)))).))))).)))))) ( -58.30) >DroSec_CAF1 3231 120 - 1 GUUGCUGGAGGAGCAGCGGCGACUCCAGCGGGAACUGGUCGAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUCAACUGCGCCCUCCGCGGAGUGCUAC .((((.((((((((((((((..(((((((((((.((..((....))..)).)))))))))))..)))))))))((((((....((((....)))).)))).)))))))))))........ ( -62.30) >DroSim_CAF1 3534 120 - 1 GUUGCUGGAGGAGCAGCGGCGACUCCAGCGGGAACUGGUCGAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUCAACUGCGCCCUCCGCGGAGUGCUAC .((((.((((((((((((((..(((((((((((.((..((....))..)).)))))))))))..)))))))))((((((....((((....)))).)))).)))))))))))........ ( -62.30) >DroPer_CAF1 12117 120 - 1 GUUGCAGCAGCAGCAGCGGCGACUCCACCGGAUACUGAUCGAUGGAGAGGCUCUUGUUCGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUCAGCUGGGCGCGACGCGGUGUGCUCC ....((.(.(((((((((((..(((((.(((.......))).)))))..((((......)))).))))))))))).).))..(((((((((((((.((.....)))).))))).)))))) ( -58.30) >consensus GUUGCUGGAGGAGCAGCGGCGACUCCACCGGGAACUGGUCGAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUCAGCUGCGCCCUCCGCGGAGUGCUAC .((((.((((((((((((((..(((((.(((.......))).)))))..((((......)))).)))))))))((((((....((((....)))).)))).)))))))))))........ (-52.58 = -52.70 + 0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:08 2006