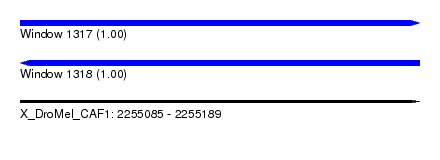
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,255,085 – 2,255,189 |
| Length | 104 |
| Max. P | 0.999997 |

| Location | 2,255,085 – 2,255,189 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 67.84 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -12.85 |
| Energy contribution | -15.50 |
| Covariance contribution | 2.65 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
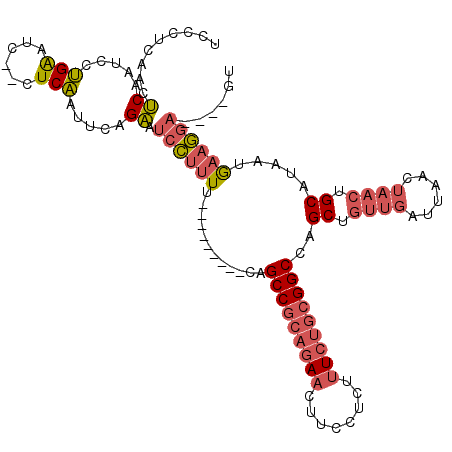
>X_DroMel_CAF1 2255085 104 + 22224390 UCCCUCAACUCAAUCCUGAAUC--CUCAAUUCAGAAUCCUUUU---------CAGCCGCAGAACUUCCUCUUUCUGCGGCCAGCUGUUGAUUAACUAACUGCAUAAUGAAGGA----GU .......((((....((((((.--....))))))......(((---------(((((((((((........)))))))))..((.((((......)))).))....)))))))----)) ( -30.40) >DroGri_CAF1 24785 105 + 1 UCAAUCU-GGCAGUACGGGUUCUUUUCCAUUUAGCGUCGCCUCGAGCUGUGGCAGCC-----ACUUCCUUU--CUGCGGCCAGCUGUUGAUUAA----GUGCAUAAUAACUACACAC-- ..((((.-((((((..(((......))).....(.(((((...(((..((((...))-----))...))).--..)))))).))))))))))..----(((...........)))..-- ( -23.50) >DroSec_CAF1 17933 104 + 1 UCCCUCAACUCAAUCCUGAAUC--CUCAAUUCAGAAUCCUUUU---------CAGCCGCAGAACUUCCUCUUUCUGCGGCCAGCUGUUGAUUAACUAACUGCAUAAUGAAGGA----GU .......((((....((((((.--....))))))......(((---------(((((((((((........)))))))))..((.((((......)))).))....)))))))----)) ( -30.40) >consensus UCCCUCAACUCAAUCCUGAAUC__CUCAAUUCAGAAUCCUUUU_________CAGCCGCAGAACUUCCUCUUUCUGCGGCCAGCUGUUGAUUAACUAACUGCAUAAUGAAGGA____GU .........((.....(((......))).....)).((((((............(((((((((........)))))))))..((.((((......)))).)).....))))))...... (-12.85 = -15.50 + 2.65)

| Location | 2,255,085 – 2,255,189 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 67.84 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -20.92 |
| Energy contribution | -23.23 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.61 |
| SVM decision value | 6.16 |
| SVM RNA-class probability | 0.999997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
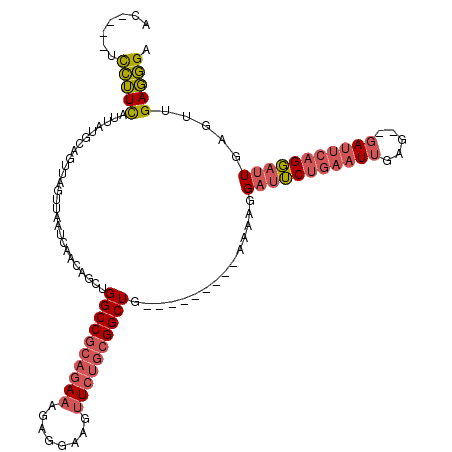
>X_DroMel_CAF1 2255085 104 - 22224390 AC----UCCUUCAUUAUGCAGUUAGUUAAUCAACAGCUGGCCGCAGAAAGAGGAAGUUCUGCGGCUG---------AAAAGGAUUCUGAAUUGAG--GAUUCAGGAUUGAGUUGAGGGA ..----(((((((.....(((((.(((....))))))))(((((((((........)))))))))..---------.....(((((((((((...--)))))))))))....))))))) ( -38.10) >DroGri_CAF1 24785 105 - 1 --GUGUGUAGUUAUUAUGCAC----UUAAUCAACAGCUGGCCGCAG--AAAGGAAGU-----GGCUGCCACAGCUCGAGGCGACGCUAAAUGGAAAAGAACCCGUACUGCC-AGAUUGA --((((((((...))))))))----........((((((((.....--......(((-----(..((((.(.....).)))).))))..((((........))))...)))-)).))). ( -26.10) >DroSec_CAF1 17933 104 - 1 AC----UCCUUCAUUAUGCAGUUAGUUAAUCAACAGCUGGCCGCAGAAAGAGGAAGUUCUGCGGCUG---------AAAAGGAUUCUGAAUUGAG--GAUUCAGGAUUGAGUUGAGGGA ..----(((((((.....(((((.(((....))))))))(((((((((........)))))))))..---------.....(((((((((((...--)))))))))))....))))))) ( -38.10) >consensus AC____UCCUUCAUUAUGCAGUUAGUUAAUCAACAGCUGGCCGCAGAAAGAGGAAGUUCUGCGGCUG_________AAAAGGAUUCUGAAUUGAG__GAUUCAGGAUUGAGUUGAGGGA .......(((((..........................((((((((((........))))))))))...............(((((((((((.....))))))))))).....))))). (-20.92 = -23.23 + 2.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:26 2006