
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,263,773 – 20,263,864 |
| Length | 91 |
| Max. P | 0.505895 |

| Location | 20,263,773 – 20,263,864 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.74 |
| Mean single sequence MFE | -35.25 |
| Consensus MFE | -23.10 |
| Energy contribution | -23.72 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
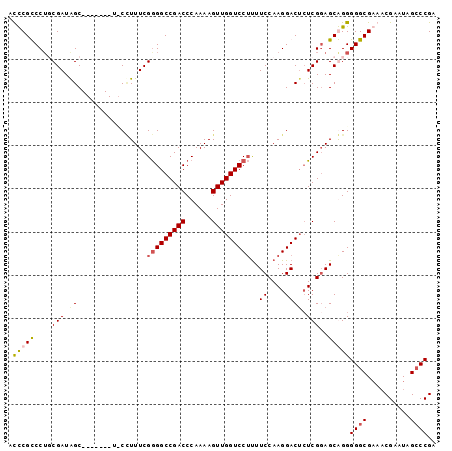
>X_DroMel_CAF1 20263773 91 + 22224390 AUCCGCCCUGCGAUGGC-------U-CCUUUCGGGGCCGACCCAAAAGUUGGUCCUUUUCCAAGGACUCUCGGAGCGGGGGGCGAAACGAAUAGACCGA ...((((((.((.((((-------(-((....)))))))........(..(((((((....)))))))..)....)).))))))............... ( -36.00) >DroSec_CAF1 10451 91 + 1 AUCCGCCCAGCGAUAGC-------U-CCUUUCGGGGCCGACCCAAAAGUUGGUCCUUUUCCAAGGACUCUCGGAGCGGGGGGCGCAACGAGUGGCCCGA ..((((((.((....))-------(-((((..(((((((((......))))))))).....))))).....)).))))(((.(((.....))).))).. ( -39.20) >DroEre_CAF1 10240 99 + 1 ACUCGUCCUUCGAUAGCCGAUGGCCUCUUUUCGAGGCCGACCCAAAAGUUGGUCCACUUCCAAGGACAAUCGGAGCAGGGGGCGAAACGAAUAGCCCGA ..(((((((..(....(((((((((((.....)))))).............((((........)))).)))))..)..))))))).............. ( -36.80) >DroYak_CAF1 10655 92 + 1 ACUCGUCCUUCGAUAGC-------UCCAUUUCGGGGCCGACCCAAAAGUUGGUCCUUUUCCAAGGACUCUCGUAGCAGGGGGCGAACCGAAUAGCCCGA ......(((.(((.((.-------(((.....(((((((((......))))))))).......))))).)))....)))((((..........)))).. ( -29.00) >consensus ACCCGCCCUGCGAUAGC_______U_CCUUUCGGGGCCGACCCAAAAGUUGGUCCUUUUCCAAGGACUCUCGGAGCAGGGGGCGAAACGAAUAGCCCGA .(((((....(((.((................(((((((((......)))))))))..((....)))).)))..)))))((((..........)))).. (-23.10 = -23.72 + 0.62)


| Location | 20,263,773 – 20,263,864 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.74 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -23.60 |
| Energy contribution | -25.22 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
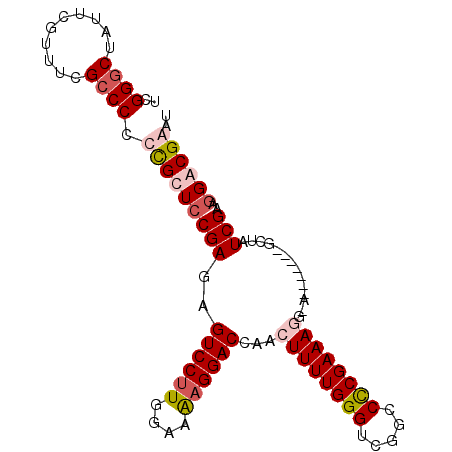
>X_DroMel_CAF1 20263773 91 - 22224390 UCGGUCUAUUCGUUUCGCCCCCCGCUCCGAGAGUCCUUGGAAAAGGACCAACUUUUGGGUCGGCCCCGAAAGG-A-------GCCAUCGCAGGGCGGAU .(((.....))).(((((((...(((((((((((((((....))))))....)))))))..(((.((....))-.-------)))...)).))))))). ( -38.20) >DroSec_CAF1 10451 91 - 1 UCGGGCCACUCGUUGCGCCCCCCGCUCCGAGAGUCCUUGGAAAAGGACCAACUUUUGGGUCGGCCCCGAAAGG-A-------GCUAUCGCUGGGCGGAU ..((((((.....)).)))).((((((((((.((((((....)))))))..((((((((.....)))))))).-.-------....)))..)))))).. ( -39.40) >DroEre_CAF1 10240 99 - 1 UCGGGCUAUUCGUUUCGCCCCCUGCUCCGAUUGUCCUUGGAAGUGGACCAACUUUUGGGUCGGCCUCGAAAAGAGGCCAUCGGCUAUCGAAGGACGAGU ((((((.................)).))))((((((((.(((((.((((........))))((((((.....))))))....))).)).)))))))).. ( -39.83) >DroYak_CAF1 10655 92 - 1 UCGGGCUAUUCGGUUCGCCCCCUGCUACGAGAGUCCUUGGAAAAGGACCAACUUUUGGGUCGGCCCCGAAAUGGA-------GCUAUCGAAGGACGAGU (((((((.(((((...((((((........(.((((((....))))))).......)))..))).)))))....)-------))).((....))))).. ( -29.86) >consensus UCGGGCUAUUCGUUUCGCCCCCCGCUCCGAGAGUCCUUGGAAAAGGACCAACUUUUGGGUCGGCCCCGAAAGG_A_______GCUAUCGAAGGACGAAU ..((((..........)))).(((((((((..((((((....))))))...((((((((.....))))))))..............)))..)))))).. (-23.60 = -25.22 + 1.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:54 2006