
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,251,789 – 20,251,906 |
| Length | 117 |
| Max. P | 0.705604 |

| Location | 20,251,789 – 20,251,906 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -46.99 |
| Consensus MFE | -29.79 |
| Energy contribution | -29.82 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20251789 117 + 22224390 GCUGU---GAAACGCCGCUGAUCCAGGCCAGCCAGAGCACCAGCAUCGAGCUGGUAUGCUCCACCAUGCGCCGGCAGAUCAUCUCUAGGGCCUUCUACGGCUGGCUGGCCUACUGUCGCC ((.((---(...))).)).(((..((((((((((((((((((((.....))))))..))))........((((..(((....((....))...))).))))))))))))))...)))... ( -48.90) >DroVir_CAF1 5915 120 + 1 GCAACCCAGAUCCACCAUUGAUACAGGCCAGCCAAACGGCCAGCAUUGAGCUGGUCUGCUCCACAAUGCGUCGCCAGAUAAUCUCGCGCGCCUUCUACGGCUGGCUGGCCUACUGUCGGC .................((((((.((((((((((...(((((((.....))))))).(((.......(((.(((.((.....)).)))))).......)))))))))))))..)))))). ( -48.34) >DroGri_CAF1 5382 120 + 1 GCAAUCCGGAUCCACCAUUGAUUCAGGCCAGCCAAACGGCCAGCAUCGAGCUCGUCUGCUCCACGAUGCGUCGCCAGAUCAUCUCGCGCGCCUUCUACGGCUGGCUGGCCUACUGUCGCC ((.....(((((.......)))))((((((((((...(((..(((((((((......))))...)))))...)))..............(((......)))))))))))))......)). ( -48.20) >DroWil_CAF1 1043 117 + 1 ACAAU---GAGACGCCAUUGAUACAGGCCAGUCAAACGGCCAGCAUUGAGCUUGUCUGCUCCACCAUGCGGCGUCAAAUAAUCUCGAGAGCUUUCUAUGGCUGGUUGGCCUAUUGCCGUC .....---..((((((.........((((........)))).((((.((((......))))....)))))))))).....................(((((.((....))....))))). ( -37.10) >DroAna_CAF1 9985 117 + 1 CCUGC---GAGACGCCGCUGAUGCAGGCCAGCCAGAGCACGAGCAUCGAGCUGGUCUGCUCCACGAUGAGGCGCCAGAUCAUCUCCCGGGCGUUUUACGGGUGGCUGGCCUACUGCCGCC ((((.---.((((((((.((..((((((((((..((((....)).))..))))))))))..)))(((((..(....).))))).....)))))))..)))).(((.(((.....)))))) ( -55.20) >DroPer_CAF1 1672 117 + 1 GCAGC---GAUCCGCCCUUGAUCCAGGCCAGUCAGACGGCCAGCAUUGAGCUCGUGUGCUCCACCAUGCGCCGGCAGAUCAUCUCGCGCGCCUUCUACGGCUGGCUGGCCUAUUGCCGUC ((((.---((((.......)))).((((((((((..((((..((((.((((......))))....))))))))((((.....)).))..(((......))))))))))))).)))).... ( -44.20) >consensus GCAAC___GAUACGCCAUUGAUACAGGCCAGCCAAACGGCCAGCAUCGAGCUGGUCUGCUCCACCAUGCGCCGCCAGAUCAUCUCGCGCGCCUUCUACGGCUGGCUGGCCUACUGCCGCC ........................((((((((((........((((.((((......))))....))))....................(((......)))))))))))))......... (-29.79 = -29.82 + 0.03)
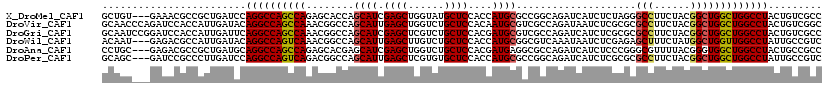
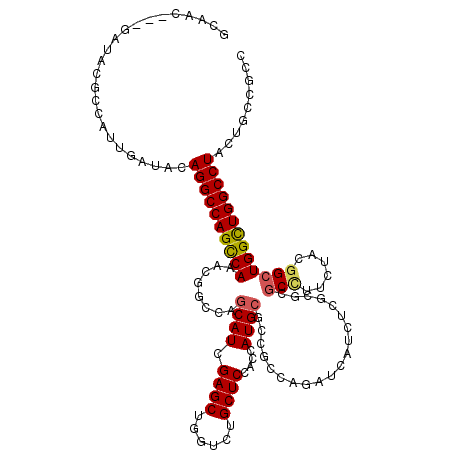
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:49 2006