
| Sequence ID | X_DroMel_CAF1 |
|---|---|
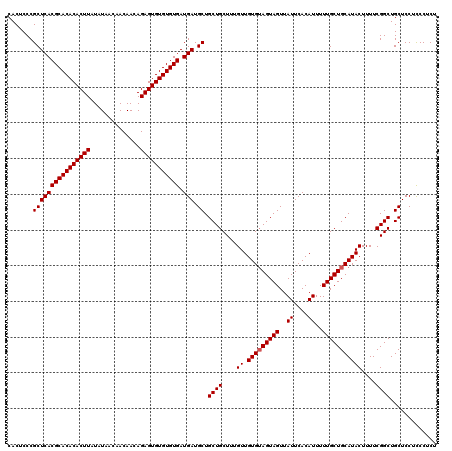
| Location | 20,244,767 – 20,244,923 |
| Length | 156 |
| Max. P | 0.991407 |

| Location | 20,244,767 – 20,244,887 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -33.61 |
| Consensus MFE | -32.34 |
| Energy contribution | -32.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20244767 120 - 22224390 CACUCCCGCUCACGCACACACUUAUAUAACAACAACAGAGUGUGUGUGAUGAUGCUGCUGCUUUGUUGUGUAGUAGUUAUUCACAUUUUUGCUGCAUACUUUUCGGCUGCUCCUCCCCCU .......((((((((((((((((..............))))))))))).))).)).((((....((.(((((((((..((....))..)))))))))))....))))............. ( -33.84) >DroSim_CAF1 362 120 - 1 CACUCCCGCUCACGCACACACUUAUAUAACAACAACAGAGUGUGUGUGAUGAUGCUGCUGCUUUGUUGUGUAGUAGUUAUUCACAUUUUUGCUGCAUACUUUUCGGCUGCUCCUCCCUCU .......((((((((((((((((..............))))))))))).))).)).((((....((.(((((((((..((....))..)))))))))))....))))............. ( -33.84) >DroYak_CAF1 1550 120 - 1 CACUCCCGCUCACGCACACACUUAUAUAACAACAACAGAGUGUGUGUGAUGAUGCUGCUGCUUUGUUGUGAAGUAGUUAUUCACAUUUUUGCUGCAUACUUUUCGGCUGCUCCUUCCUCU .....((((((((((((((((((..............))))))))))).))).))(((.((.....((((((.......)))))).....)).)))........)).............. ( -33.14) >consensus CACUCCCGCUCACGCACACACUUAUAUAACAACAACAGAGUGUGUGUGAUGAUGCUGCUGCUUUGUUGUGUAGUAGUUAUUCACAUUUUUGCUGCAUACUUUUCGGCUGCUCCUCCCUCU .......((((((((((((((((..............))))))))))).))).)).((((....((.(((((((((..((....))..)))))))))))....))))............. (-32.34 = -32.67 + 0.33)


| Location | 20,244,807 – 20,244,923 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.10 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -28.67 |
| Energy contribution | -29.33 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
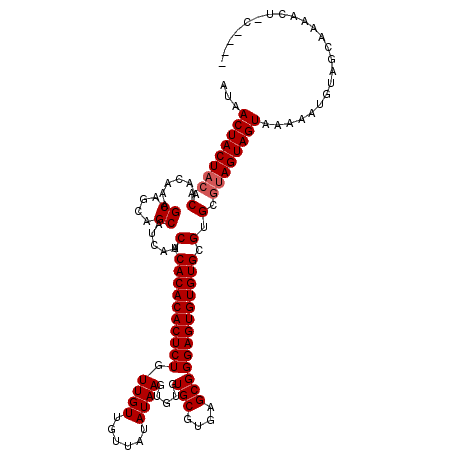
>X_DroMel_CAF1 20244807 116 + 22224390 AUAACUACUACACAACAAAGCAGCAGCAUCAUCACACACACUCUGUUGUUGUUAUAUAAGUGUGUGCGUGAGCGGGAGUGUGUGCGUGCGUAGUAGUAAAAAUGUAGCAAAACUGC---- ...((((((((.((.....((....)).....(.((((((((((.((((......)))).....(((....))))))))))))).))).))))))))......((((.....))))---- ( -35.30) >DroSim_CAF1 402 113 + 1 AUAACUACUACACAACAAAGCAGCAGCAUCAUCACACACACUCUGUUGUUGUUAUAUAAGUGUGUGCGUGAGCGGGAGUGUGUGCGCGCAUAGUAGUAAAAAUGUAGCAAAA---C---- .....((((((.......((((((((((...............)))))))))).......((((((((((.((....))...))))))))))))))))..............---.---- ( -31.86) >DroYak_CAF1 1590 120 + 1 AUAACUACUUCACAACAAAGCAGCAGCAUCAUCACACACACUCUGUUGUUGUUAUAUAAGUGUGUGCGUGAGCGGGAGUGUGUGCGUGCGUAGUAGUAAAAAUGUAGCAAAACUACAAUU ...((((((.(((......((....)).......((((((((((.((((......)))).....(((....))))))))))))).)))...)))))).....(((((.....)))))... ( -34.90) >consensus AUAACUACUACACAACAAAGCAGCAGCAUCAUCACACACACUCUGUUGUUGUUAUAUAAGUGUGUGCGUGAGCGGGAGUGUGUGCGUGCGUAGUAGUAAAAAUGUAGCAAAACU_C____ ...((((((((.(......((....)).....(.(((((((((((((..(((.((((....)))))))..))).)))))))))).).).))))))))....................... (-28.67 = -29.33 + 0.67)

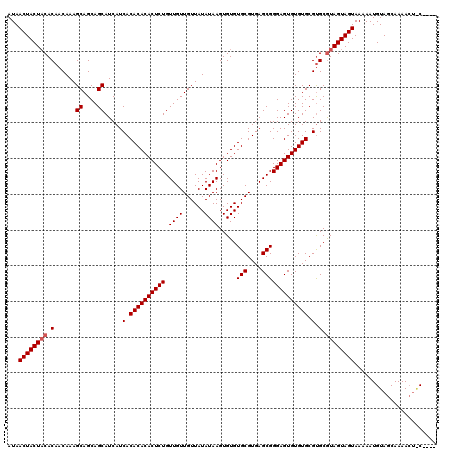
| Location | 20,244,807 – 20,244,923 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.10 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -32.78 |
| Energy contribution | -32.67 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20244807 116 - 22224390 ----GCAGUUUUGCUACAUUUUUACUACUACGCACGCACACACUCCCGCUCACGCACACACUUAUAUAACAACAACAGAGUGUGUGUGAUGAUGCUGCUGCUUUGUUGUGUAGUAGUUAU ----((......)).........(((((((((((.(((....(..(.((((((((((((((((..............))))))))))).))).)).)..)...))))))))))))))... ( -35.94) >DroSim_CAF1 402 113 - 1 ----G---UUUUGCUACAUUUUUACUACUAUGCGCGCACACACUCCCGCUCACGCACACACUUAUAUAACAACAACAGAGUGUGUGUGAUGAUGCUGCUGCUUUGUUGUGUAGUAGUUAU ----.---..(((((((((....((......(((.(((.........((((((((((((((((..............))))))))))).))).))))))))...)).))))))))).... ( -31.64) >DroYak_CAF1 1590 120 - 1 AAUUGUAGUUUUGCUACAUUUUUACUACUACGCACGCACACACUCCCGCUCACGCACACACUUAUAUAACAACAACAGAGUGUGUGUGAUGAUGCUGCUGCUUUGUUGUGAAGUAGUUAU ...(((((.....))))).....((((((.((((.(((....(..(.((((((((((((((((..............))))))))))).))).)).)..)...))))))).))))))... ( -34.94) >consensus ____G_AGUUUUGCUACAUUUUUACUACUACGCACGCACACACUCCCGCUCACGCACACACUUAUAUAACAACAACAGAGUGUGUGUGAUGAUGCUGCUGCUUUGUUGUGUAGUAGUUAU .......................(((((((((((.(((....(..(.((((((((((((((((..............))))))))))).))).)).)..)...))))))))))))))... (-32.78 = -32.67 + -0.11)


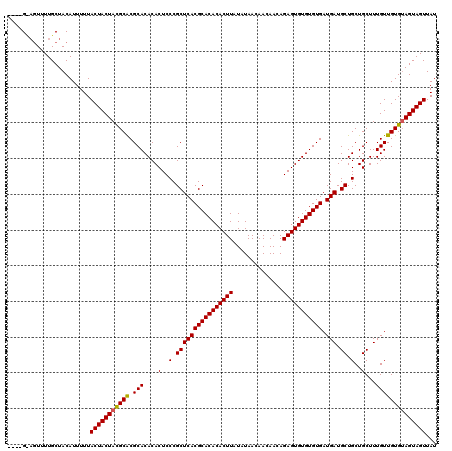
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:42 2006