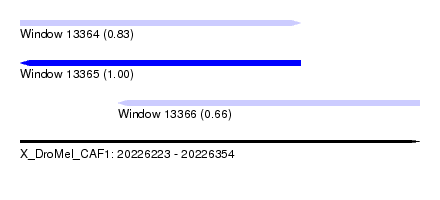
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,226,223 – 20,226,354 |
| Length | 131 |
| Max. P | 0.998729 |

| Location | 20,226,223 – 20,226,315 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -27.55 |
| Energy contribution | -27.93 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20226223 92 + 22224390 AGCCCGGCCAGAAAGUAAUCCGCCCCAGGAACACCACCCACCGCCC-----------------GCCCAUUUGGAUAUGGAAAUGGGCAGUGGGGGCGGCGAUUGGCGCU (((...((((((......))((((((.((....)).(((((.((((-----------------(.((((......))))...))))).))))))).))))..))))))) ( -38.10) >DroSec_CAF1 13225 85 + 1 -------CCAGAAAGCAGUCCGCCCCAGGAACACCGCCCACUGCCC-----------------GCCCAUUUGGAUAUGGAAAUGGGCAGUGGGGGCGGCGAUUGGCGCU -------......(((.(((((((((.((....)).((((((((((-----------------(.((((......))))...))))))))))))).))))...)))))) ( -40.90) >DroSim_CAF1 12532 91 + 1 AGCCCGGCCAGAAAGCAGUCCGCCCCAGGAACACCGCCCACUGCCC-----------------GCCCAUUUGGAUAUGGAAAUGGGCAGUGG-GGCGGCGAUUGGCGCU (((...(((((.......(((......))).(.(((((((((((((-----------------(.((((......))))...)))))))).)-))))).).)))))))) ( -42.50) >DroEre_CAF1 13084 108 + 1 AGUCCGGGCAGCAAGUGAUCCGC-CCAGGCGCACCGCCCACCAGCCACCACACACCACACAGCUCCUAUUUCGAUAUGGAAAUGGGCAGUGGGGGCGGGGAUUGGCGCU .....((((............))-)).(((((.((((((.((.(......)..........(((((((((((......)))))))).)))))))))))......))))) ( -40.20) >consensus AGCCCGGCCAGAAAGCAAUCCGCCCCAGGAACACCGCCCACCGCCC_________________GCCCAUUUGGAUAUGGAAAUGGGCAGUGGGGGCGGCGAUUGGCGCU .....................((.((((...(.((((((.((((...................((((((((........)))))))).)))))))))).).)))).)). (-27.55 = -27.93 + 0.38)

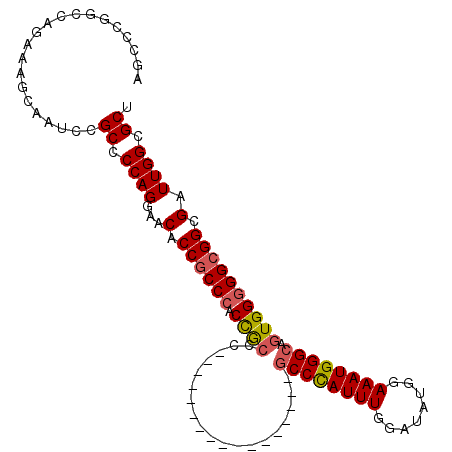

| Location | 20,226,223 – 20,226,315 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -46.97 |
| Consensus MFE | -31.25 |
| Energy contribution | -31.38 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.67 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20226223 92 - 22224390 AGCGCCAAUCGCCGCCCCCACUGCCCAUUUCCAUAUCCAAAUGGGC-----------------GGGCGGUGGGUGGUGUUCCUGGGGCGGAUUACUUUCUGGCCGGGCU (((((((..((((((((.....((((((((........))))))))-----------------))))))))..))))))).((.((.((((......)))).)).)).. ( -48.00) >DroSec_CAF1 13225 85 - 1 AGCGCCAAUCGCCGCCCCCACUGCCCAUUUCCAUAUCCAAAUGGGC-----------------GGGCAGUGGGCGGUGUUCCUGGGGCGGACUGCUUUCUGG------- .((.(((..((((((((..(((((((...(((((......))))).-----------------)))))))))))))))....))).))..............------- ( -41.80) >DroSim_CAF1 12532 91 - 1 AGCGCCAAUCGCCGCC-CCACUGCCCAUUUCCAUAUCCAAAUGGGC-----------------GGGCAGUGGGCGGUGUUCCUGGGGCGGACUGCUUUCUGGCCGGGCU ...(((...(((((((-(.(((((((...(((((......))))).-----------------)))))))))))))))......((.((((......)))).)).))). ( -47.80) >DroEre_CAF1 13084 108 - 1 AGCGCCAAUCCCCGCCCCCACUGCCCAUUUCCAUAUCGAAAUAGGAGCUGUGUGGUGUGUGGUGGCUGGUGGGCGGUGCGCCUGG-GCGGAUCACUUGCUGCCCGGACU .((((......((((((((((..(((((..((((((.(.........).)))))).))).))..).))).)))))).))))((((-((((........))))))))... ( -50.30) >consensus AGCGCCAAUCGCCGCCCCCACUGCCCAUUUCCAUAUCCAAAUGGGC_________________GGGCAGUGGGCGGUGUUCCUGGGGCGGACUACUUUCUGGCCGGGCU .((.(((..(((((((..((((((((...(((((......)))))..................)))))))))))))))....))).))..................... (-31.25 = -31.38 + 0.12)


| Location | 20,226,255 – 20,226,354 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 66.44 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -14.68 |
| Energy contribution | -16.32 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20226255 99 - 22224390 -UUCGAUCGAACCGCUAAGUGGGUGUGGGAAUUAUGGGUUAGCGCCAAUCGCCGCCCCC--ACUGCCCAUUUCCAUAUCCAAAUGGGC-----------------GGGCGGUGGGUGGU -..(.(((..((((((.((((((.((((((....((((....).))).)).)))).)))--)))((((((((........))))))))-----------------.)))))).))).). ( -42.50) >DroPse_CAF1 12218 82 - 1 GCUCCAGCAAGUAUGCAAGAGGGUGUGCAAAUUACGGGGCACAGU------------GUAAACCGAUUACUUCCUGAUCU------------------------UUGUUGGUUGGCUG- ((((((((((((((((......))))))......(((((...(((------------(.........)))))))))....------------------------)))))))..)))..- ( -21.50) >DroSec_CAF1 13250 86 - 1 --------------CUAAGUGGGUGUGGGAAUUAUGGGUUAGCGCCAAUCGCCGCCCCC--ACUGCCCAUUUCCAUAUCCAAAUGGGC-----------------GGGCAGUGGGCGGU --------------.......((((..((..(((.....)))..))...))))((((((--(((((((...(((((......))))).-----------------)))))))))).))) ( -38.50) >DroSim_CAF1 12564 98 - 1 -UUCGAUCGAACCGCUAAGUGGGUGUGGGAAUUAUGGGUUAGCGCCAAUCGCCGCC-CC--ACUGCCCAUUUCCAUAUCCAAAUGGGC-----------------GGGCAGUGGGCGGU -.........((((((....(((((.(.((....((((....).))).)).)))))-)(--(((((((...(((((......))))).-----------------)))))))))))))) ( -40.70) >DroEre_CAF1 13115 116 - 1 -UUCGAUCGAACCGCUAAGUGGGUGUGGGAAUUAUGGGUUAGCGCCAAUCCCCGCCCCC--ACUGCCCAUUUCCAUAUCGAAAUAGGAGCUGUGUGGUGUGUGGUGGCUGGUGGGCGGU -.........(((((.....(((((.((((....((((....).))).)))))))))((--(((..(((((..(((((((..((((...)))).))))))).)))))..)))))))))) ( -43.50) >DroYak_CAF1 12739 103 - 1 -UUCGAUUGAAACCCUAAGAGGGUGCUGGAAUUAUGGGUUAUCGCCAAUCC-UGCCCCCAAACCGCCCAUUUCCAUAUCAAAAUGGGAG--------------GUGGCUGGCGGGCGGU -...(((((..((((.....))))((...(((.....)))...)))))))(-((((((((.(((.(((((((........))))))).)--------------))...))).)))))). ( -37.80) >consensus _UUCGAUCGAACCGCUAAGUGGGUGUGGGAAUUAUGGGUUAGCGCCAAUCGCCGCCCCC__ACUGCCCAUUUCCAUAUCCAAAUGGGC_________________GGGCGGUGGGCGGU .....................(((((...(((.....))).))))).....((((((....(((((((.....................................))))))))))))). (-14.68 = -16.32 + 1.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:35 2006