
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,224,916 – 20,225,007 |
| Length | 91 |
| Max. P | 0.992594 |

| Location | 20,224,916 – 20,225,007 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 66.48 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -13.26 |
| Energy contribution | -13.56 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20224916 91 + 22224390 CGGGUUUCCCAACUAGAGAGAAUGCUGAA-------GUCUAGUACCGACUAAUGGGAUACCCAUUAAUUACUGCUUAAAUACUGUGAUGAAAAUUGAG .((((.(((((..(((........))).(-------(((.......))))..))))).))))....(((((............))))).......... ( -19.70) >DroSec_CAF1 11755 75 + 1 CGGGUAUUCCAACUAG---------UCUA-------GUCUAGUGCCGACUAAUGGGAUACCCAUUAAUU-------AAGCAAUACGACGAAAAUUGAG .((((((((((..(((---------((..-------..........))))).)))))))))).......-------...................... ( -20.40) >DroSim_CAF1 10991 75 + 1 CGGGUAUCCCAACUAG---------UAUA-------GUCUAGUGCCGACUAAUGGGAUACCCAUUAACU-------AAGCAAUACGACGAAAAUUGAG .(((((((((((((((---------....-------..))))).........)))))))))).......-------...................... ( -22.30) >DroEre_CAF1 11859 70 + 1 CGGGUAUCCCAAAUGGAGUGAACGCUGUA-------GUCUCGUAUCGGCUG-CGGGAUACCUAUUAAU--------------------GGAAAUUGAG .(((((((((......(((....)))(((-------(((.......)))))-)))))))))).(((((--------------------....))))). ( -26.70) >DroYak_CAF1 11437 95 + 1 CGGGUAUUUCAACUGUGGAGAACGCUAUAGUCUAUAGUCUAGUAUCGACUA-CAGGACACCCACUAAUAA--GCUUAAGCAAUACAACGAAAAUUGAG .((((......(((((((......))))))).....((((.(((.....))-).))))))))........--((....)).................. ( -19.40) >consensus CGGGUAUCCCAACUAG_G_GAA_GCUAUA_______GUCUAGUACCGACUAAUGGGAUACCCAUUAAUU_______AAGCAAUACGACGAAAAUUGAG .(((((((((......((......))..........(((.......)))....))))))))).................................... (-13.26 = -13.56 + 0.30)

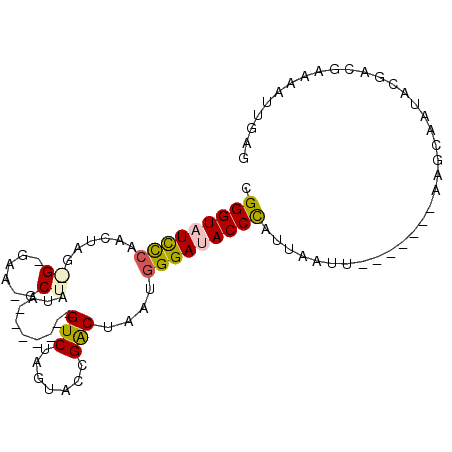
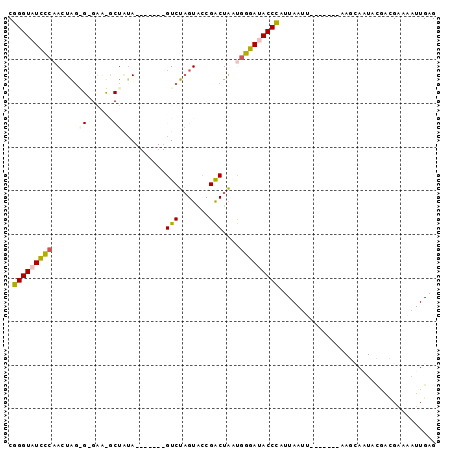
| Location | 20,224,916 – 20,225,007 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 66.48 |
| Mean single sequence MFE | -18.94 |
| Consensus MFE | -11.55 |
| Energy contribution | -12.23 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20224916 91 - 22224390 CUCAAUUUUCAUCACAGUAUUUAAGCAGUAAUUAAUGGGUAUCCCAUUAGUCGGUACUAGAC-------UUCAGCAUUCUCUCUAGUUGGGAAACCCG ....................................((((.(((((.........((((((.-------............))))))))))).)))). ( -17.62) >DroSec_CAF1 11755 75 - 1 CUCAAUUUUCGUCGUAUUGCUU-------AAUUAAUGGGUAUCCCAUUAGUCGGCACUAGAC-------UAGA---------CUAGUUGGAAUACCCG ......................-------.......((((((((.(((((((..........-------..))---------))))).)).)))))). ( -19.20) >DroSim_CAF1 10991 75 - 1 CUCAAUUUUCGUCGUAUUGCUU-------AGUUAAUGGGUAUCCCAUUAGUCGGCACUAGAC-------UAUA---------CUAGUUGGGAUACCCG ......................-------.......((((((((((.(((((.......)))-------))..---------.....)))))))))). ( -23.00) >DroEre_CAF1 11859 70 - 1 CUCAAUUUCC--------------------AUUAAUAGGUAUCCCG-CAGCCGAUACGAGAC-------UACAGCGUUCACUCCAUUUGGGAUACCCG ..........--------------------.......(((((((((-........(((....-------.....)))..........))))))))).. ( -13.97) >DroYak_CAF1 11437 95 - 1 CUCAAUUUUCGUUGUAUUGCUUAAGC--UUAUUAGUGGGUGUCCUG-UAGUCGAUACUAGACUAUAGACUAUAGCGUUCUCCACAGUUGAAAUACCCG .((((((...........((....))--......(((((...((((-(((((.(((......))).)))))))).)...)))))))))))........ ( -20.90) >consensus CUCAAUUUUCGUCGUAUUGCUU_______AAUUAAUGGGUAUCCCAUUAGUCGGUACUAGAC_______UACAGC_UUC_C_CUAGUUGGGAUACCCG ....................................((((((((((...(((.......))).........................)))))))))). (-11.55 = -12.23 + 0.68)

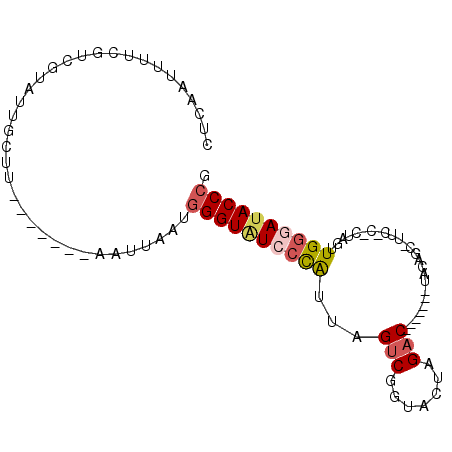
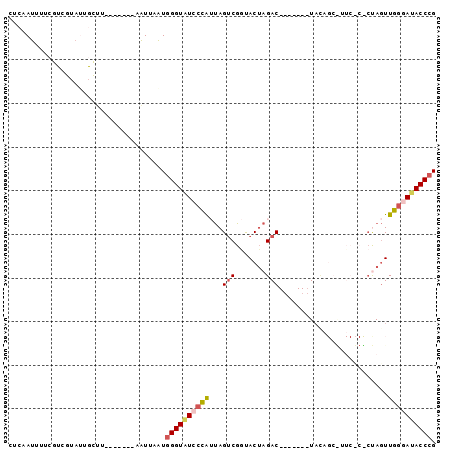
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:32 2006