| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,253,852 – 2,253,959 |
| Length | 107 |
| Max. P | 0.999794 |

| Location | 2,253,852 – 2,253,959 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.18 |
| Mean single sequence MFE | -38.23 |
| Consensus MFE | -29.23 |
| Energy contribution | -28.90 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.76 |
| SVM decision value | 4.09 |
| SVM RNA-class probability | 0.999794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2253852 107 + 22224390 -AAUUUGCAUGCCCUGUGUGCCUUUUGACCUGCGGCAACAGACGGGGCACGUGGCACUCCGAGG-AAAUG---GCAUAUGGCAUGCAGUAUAUAGCACACAGCAUAUAGCAC-------- -...(((((((((..(((((((.(((..(((..(....)...(((((.........))))))))-))).)---)))))))))))))))(((((.((.....)))))))....-------- ( -36.30) >DroSec_CAF1 16682 91 + 1 -AAUUUGCAUGCCCUGUGUGCCUUUUGACCUGCGGCAACAGACGGGGCACGUGGCACUGCGAGG-AAGCGGCGGCAUAUGGCAUGCAGCAAAU--------------------------- -...(((((((((.((((((((...((.((((((....).).)))).))...))).((((....-..))))..))))).))))))))).....--------------------------- ( -37.20) >DroYak_CAF1 17754 117 + 1 AAAUUUGCAUGACUUGUGCGCCUUUUGACCUGCGGCAACAGACGGGGCACGUGGCAGUGUAUCGCACAUG---GCAUAUGGCAUGCAGCAUGUAGCAUGUAGCAUGCAGCAAAAAGCAUA ...((((((((...((((((........((((((....).).))))((((......))))..))))))..---.)))...((((((.(((((...))))).)))))).)))))....... ( -41.20) >consensus _AAUUUGCAUGCCCUGUGUGCCUUUUGACCUGCGGCAACAGACGGGGCACGUGGCACUGCGAGG_AAAUG___GCAUAUGGCAUGCAGCAUAUAGCA___AGCAU__AGCA_________ ....(((((((((.((((((((...((.((((((....).).)))).))...))).(.....)..........))))).)))))))))................................ (-29.23 = -28.90 + -0.33)

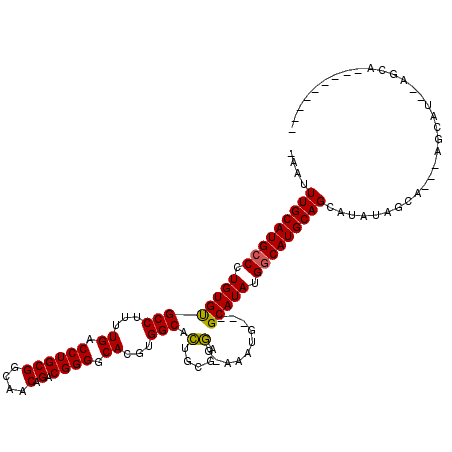
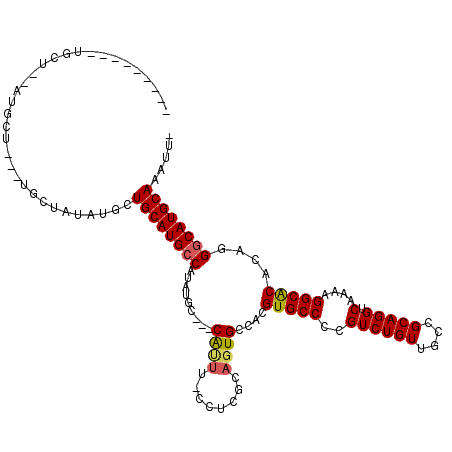
| Location | 2,253,852 – 2,253,959 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.18 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -22.82 |
| Energy contribution | -23.27 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2253852 107 - 22224390 --------GUGCUAUAUGCUGUGUGCUAUAUACUGCAUGCCAUAUGC---CAUUU-CCUCGGAGUGCCACGUGCCCCGUCUGUUGCCGCAGGUCAAAAGGCACACAGGGCAUGCAAAUU- --------(((.((((.((.....)))))))))((((((((...((.---(((((-.....))))).)).(((((..((((((....))))).)....)))))....))))))))....- ( -33.70) >DroSec_CAF1 16682 91 - 1 ---------------------------AUUUGCUGCAUGCCAUAUGCCGCCGCUU-CCUCGCAGUGCCACGUGCCCCGUCUGUUGCCGCAGGUCAAAAGGCACACAGGGCAUGCAAAUU- ---------------------------......((((((((...((.(((.((..-....)).))).)).(((((..((((((....))))).)....)))))....))))))))....- ( -31.30) >DroYak_CAF1 17754 117 - 1 UAUGCUUUUUGCUGCAUGCUACAUGCUACAUGCUGCAUGCCAUAUGC---CAUGUGCGAUACACUGCCACGUGCCCCGUCUGUUGCCGCAGGUCAAAAGGCGCACAAGUCAUGCAAAUUU .......(((((.((((((..((((...))))..))))))...(((.---(.(((((....(((......)))....((((.(((((...)).))).))))))))).).))))))))... ( -33.50) >consensus _________UGCU__AUGCU___UGCUAUAUGCUGCAUGCCAUAUGC___CAUUU_CCUCGCAGUGCCACGUGCCCCGUCUGUUGCCGCAGGUCAAAAGGCACACAGGGCAUGCAAAUU_ .................................((((((((.........((((........))))....(((((..((((((....))))).)....)))))....))))))))..... (-22.82 = -23.27 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:23 2006