| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,222,136 – 20,222,246 |
| Length | 110 |
| Max. P | 0.634849 |

| Location | 20,222,136 – 20,222,246 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.00 |
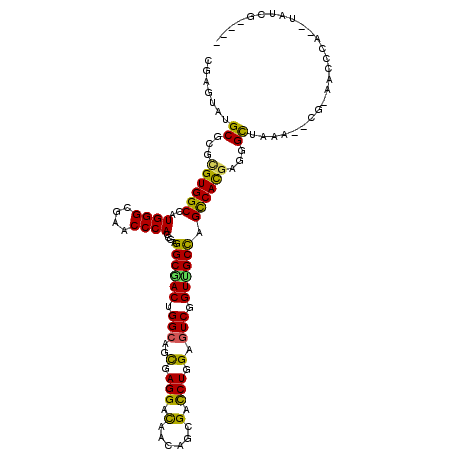
| Mean single sequence MFE | -40.73 |
| Consensus MFE | -28.86 |
| Energy contribution | -27.67 |
| Covariance contribution | -1.19 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20222136 110 + 22224390 CGAGUACGCACGCGUGGCCAUGGGCGAACCCACCGAGGCCACCGGCAGCGAGGACAACAGCGACCUAGAGUCGGUGGCUAGCCACGAGGGUUGAA--CCGAACCCA-CUGUCC---- ..(((.......((((((..((((....))))....((((((((((....(((.(......).)))...)))))))))).)))))).(((((...--...))))))-))....---- ( -45.00) >DroVir_CAF1 10652 110 + 1 UGAAUAUGCGCGUGUGGCCAUGGGCGAACCCACCGAGGCGACUGGCAGCGAGGACAACAGCGAUCUCGAGUCCGUUGCCAGUCACGAUGGCUGAA--CG-----CAAUUAUCGAUAA .........((((.((((((((((....)))......(.((((((((((..((((.....((....)).)))))))))))))).).))))))).)--))-----)............ ( -43.70) >DroWil_CAF1 9337 100 + 1 CGAGUAUGCUCGUGUGGCCAUGGGCGAACCCACCGAGGCAACAGGCAGUGAGGACAAUAGCGAUCUAGAGUCAGUUGCCAGUCAUGAAGGCUAAA--A--AACC------------- ((((....))))..(((((.((((....))))....((((((.(((....(((.(......).)))...))).)))))).........)))))..--.--....------------- ( -29.20) >DroMoj_CAF1 9601 109 + 1 UGAAUAUGCGCGCGUGGCCAUGGGCGAACCCACCGAGGCAACUGGGAGCGAGGACAACAGCGAUCUGGAGUCCGUUGCCAGUCACGAUGGCUAAA--CG-----CA-CUAUCGAUAU .........(((..((((((((((....))).((.((....)).)).((((((((..(((....)))..)))).))))........)))))))..--))-----).-.......... ( -39.60) >DroAna_CAF1 9435 109 + 1 UGAGUAUGCCCGGGUGGCGAUGGGCGAACCCACCGAGGCGACCGGCAGCGAGGACAACAGCGACCUGGAGUCGGUGGCCAGCCACGAGGGCUAAGACCCCAACC----UGCCU---- ...((..((((..((((((.((((....)))).)..(((.((((((..(.(((.(......).))).).)))))).))).)))))..))))....)).......----.....---- ( -45.80) >DroPer_CAF1 9564 110 + 1 CGAGUAUGCGCGCGUGGCCAUGGGCGAACCCACCGAGGCGACUGGCAGCGAGGAUAACAGCGACCUGGAGUCGGUUGCCAGCCACGACGGCUAAU--GGACACGUC-ACAACA---- ...((.((.(.(((((.(((((((....))).....((((((((((..(.(((..........))).).))))))))))((((.....)))).))--)).))))))-.)))).---- ( -41.10) >consensus CGAGUAUGCGCGCGUGGCCAUGGGCGAACCCACCGAGGCGACUGGCAGCGAGGACAACAGCGACCUGGAGUCGGUUGCCAGCCACGAGGGCUAAA__CG_AACCCA__UAUCG____ .......((...((((((..((((....))))....((((((.(((..(.(((.(......).))).).))).)))))).))))))...)).......................... (-28.86 = -27.67 + -1.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:24 2006