| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,205,584 – 20,205,742 |
| Length | 158 |
| Max. P | 0.900351 |

| Location | 20,205,584 – 20,205,677 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 70.25 |
| Mean single sequence MFE | -43.38 |
| Consensus MFE | -16.69 |
| Energy contribution | -18.09 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20205584 93 + 22224390 CGG---AUCCAGUGGCCAGGGUGCUGGGUCCAGCCAGACCGCUUCUGCAGCACCAGUCUCCGCUGG---------AGUUGGAGUAGGAGGGGGUGGAGGUGCCAG .((---((((((((.(....)))))))))))..(((..((.((((((((((.(((((....)))))---------.)))...))))))).)).)))......... ( -40.40) >DroGri_CAF1 16284 96 + 1 CGAAUAAUCCGGUGG---------CGGCUCCAGCUGCGUAACCACACCAGCAGCAGUCGCAGCAGAUGUGGCUGGAGCUGGUGCUGGUGGCGGCUGUGGUGCGAG ..............(---------((.(..(((((((...(((((((((((..((((((((.....))))))))..))))))).)))).)))))))..))))... ( -48.90) >DroSec_CAF1 33807 93 + 1 CGG---AUCCAGUGGCCAAGGUGCUGGGUCCAGCCAGCCCGCUUCUGCAGCACCAGUCUCCGCUGG---------AGUUGGUGUAGGAGGGGGUGGAGGUGCCAG .((---((((((((.(....))))))))))).(((.((((.((((((((.(((((((....)))))---------...)).)))))))).))))...)))..... ( -47.60) >DroSim_CAF1 33464 93 + 1 CGG---AUCCAGUGGCCAAGGUGCUGGGUCCAGCCAGCCCGCUUCUGCAGCACCAGUCUCCGCUGG---------AGUUGGUGUAGGAGGGGGUGGAGGUGCCAG .((---((((((((.(....))))))))))).(((.((((.((((((((.(((((((....)))))---------...)).)))))))).))))...)))..... ( -47.60) >DroEre_CAF1 22025 81 + 1 UGG---AUCCAGUGGCCAGGGUGCUGGCACCAGCCAGUCCGCUGCUGCACCACCAGUUUCCGCGGG---------AGUAGGG------------GGAGUUGCCAG (((---.(((.(..((.((.(..(((((....)))))..).))))..)....((..((((....))---------))..)).------------)))....))). ( -32.40) >consensus CGG___AUCCAGUGGCCAAGGUGCUGGGUCCAGCCAGCCCGCUUCUGCAGCACCAGUCUCCGCUGG_________AGUUGGUGUAGGAGGGGGUGGAGGUGCCAG ............((((...((.(((((......)))))))((((((((.........((((((.............)).)))).........)))))))))))). (-16.69 = -18.09 + 1.40)

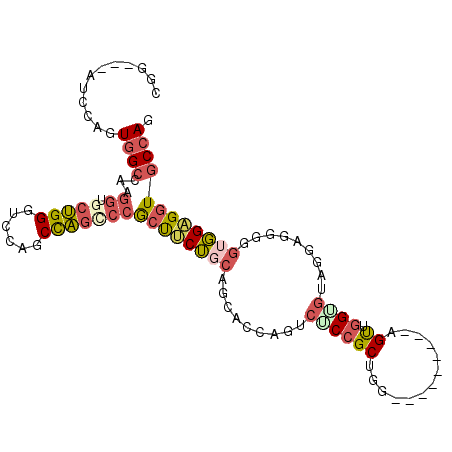

| Location | 20,205,616 – 20,205,712 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 66.96 |
| Mean single sequence MFE | -42.48 |
| Consensus MFE | -18.32 |
| Energy contribution | -20.07 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20205616 96 + 22224390 AGACCGCUUCUGCAGCACCAGUCUCCGCUGG---------AGUUGGAGUAGGAGGGGGUGGAGGUGCCAGUGGA---GCAGCGGCAGGAGCAGGCUCCGCAGCAGGAU ...((((((((.(.((.((((.((((...))---------)))))).)).)..))))))))...(((..(((((---((....((....))..))))))).))).... ( -38.70) >DroGri_CAF1 16310 94 + 1 GCGUAACCACACCAGCAGCAGUCGCAGCAGAUGUGGCUGGAGCUGGUGCUGGUGGCGGCUGUGGUGCGAGUAGCUUGGUAACGGCUUGAUGA----------CU---- .(((.(((((((((((..((((((((.....))))))))..))))))).)))).)))(((((...((.....))......))))).......----------..---- ( -42.10) >DroSec_CAF1 33839 96 + 1 AGCCCGCUUCUGCAGCACCAGUCUCCGCUGG---------AGUUGGUGUAGGAGGGGGUGGAGGUGCCAGUGGA---GCAGCGGCAGGAGCGGGCUCCGCAGCAGGAU .((((((((((((.(((((((.((((...))---------))))))))).............(.((((....).---))).).))))))))))))(((......))). ( -47.30) >DroSim_CAF1 33496 96 + 1 AGCCCGCUUCUGCAGCACCAGUCUCCGCUGG---------AGUUGGUGUAGGAGGGGGUGGAGGUGCCAGUGGA---GCAGCGGCAGGAGCAGGCUCCGCAGCAGGAU .((((.((((((((.(((((((....)))))---------...)).)))))))).)))).....(((..(((((---((....((....))..))))))).))).... ( -45.30) >DroEre_CAF1 22057 84 + 1 AGUCCGCUGCUGCACCACCAGUUUCCGCGGG---------AGUAGGG------------GGAGUUGCCAGUGUA---GCAGCGGUAGGAGCGAGCUCCGCGGCAGGAU .((((((((((((((...((..((((.(...---------....).)------------)))..))...)))))---))))).((.((((....))))...)).)))) ( -39.00) >consensus AGCCCGCUUCUGCAGCACCAGUCUCCGCUGG_________AGUUGGUGUAGGAGGGGGUGGAGGUGCCAGUGGA___GCAGCGGCAGGAGCAGGCUCCGCAGCAGGAU .(((.((((((((.((......((((((.............................)))))).(((..........))))).)))))))).)))(((......))). (-18.32 = -20.07 + 1.75)


| Location | 20,205,616 – 20,205,712 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 66.96 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -13.18 |
| Energy contribution | -14.53 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20205616 96 - 22224390 AUCCUGCUGCGGAGCCUGCUCCUGCCGCUGC---UCCACUGGCACCUCCACCCCCUCCUACUCCAACU---------CCAGCGGAGACUGGUGCUGCAGAAGCGGUCU (((((.(((((((((..((.......)).))---)))...((((((........((((..........---------.....))))...)))))))))).)).))).. ( -29.86) >DroGri_CAF1 16310 94 - 1 ----AG----------UCAUCAAGCCGUUACCAAGCUACUCGCACCACAGCCGCCACCAGCACCAGCUCCAGCCACAUCUGCUGCGACUGCUGCUGGUGUGGUUACGC ----..----------.........(((.((((.((.....))...........((((((((.(((.((((((.......)))).))))).)))))))))))).))). ( -32.20) >DroSec_CAF1 33839 96 - 1 AUCCUGCUGCGGAGCCCGCUCCUGCCGCUGC---UCCACUGGCACCUCCACCCCCUCCUACACCAACU---------CCAGCGGAGACUGGUGCUGCAGAAGCGGGCU .(((......)))(((((((.((((((((.(---(((.((((..........................---------)))).))))...))))..)))).))))))). ( -37.67) >DroSim_CAF1 33496 96 - 1 AUCCUGCUGCGGAGCCUGCUCCUGCCGCUGC---UCCACUGGCACCUCCACCCCCUCCUACACCAACU---------CCAGCGGAGACUGGUGCUGCAGAAGCGGGCU .(((......)))(((((((.((((((((.(---(((.((((..........................---------)))).))))...))))..)))).))))))). ( -35.47) >DroEre_CAF1 22057 84 - 1 AUCCUGCCGCGGAGCUCGCUCCUACCGCUGC---UACACUGGCAACUCC------------CCCUACU---------CCCGCGGAAACUGGUGGUGCAGCAGCGGACU ..........((((....))))..(((((((---(.(((((....)...------------.......---------.(((.(....)))).)))).))))))))... ( -31.80) >consensus AUCCUGCUGCGGAGCCUGCUCCUGCCGCUGC___UCCACUGGCACCUCCACCCCCUCCUACACCAACU_________CCAGCGGAGACUGGUGCUGCAGAAGCGGGCU .(((......)))..(((((.((((.((......(((.((((...................................)))).))).......)).)))).)))))... (-13.18 = -14.53 + 1.35)

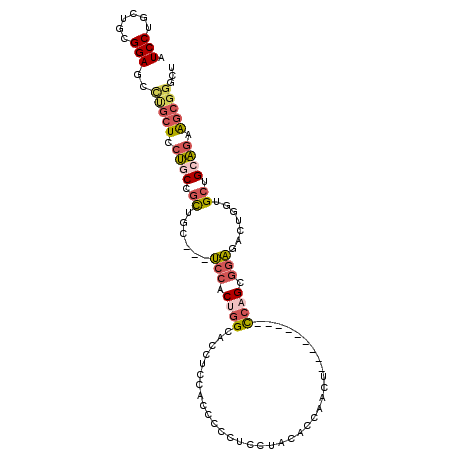

| Location | 20,205,647 – 20,205,742 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 84.69 |
| Mean single sequence MFE | -35.15 |
| Consensus MFE | -28.49 |
| Energy contribution | -29.05 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20205647 95 + 22224390 AGUUGGAGUAGGAGGGGGUGGAGGUGCCAGUGGAGCAGCGGCAGGAGCAGGCUCCGCAGCAGGAUCUGGAUCGG------GAUCAGGUUCCGGAUCAGCAG ................(((......))).(((((((....((....))..))))))).((..((((((((((..------.....)).)))))))).)).. ( -32.50) >DroSec_CAF1 33870 95 + 1 AGUUGGUGUAGGAGGGGGUGGAGGUGCCAGUGGAGCAGCGGCAGGAGCGGGCUCCGCAGCAGGAUCUGGAUCCG------GAUCAGGUUCCGGAUCAGCAG .((((((...(((...(((......))).(((((((.((.......))..))))))).((..((((((....))------))))..)))))..)))))).. ( -38.20) >DroSim_CAF1 33527 95 + 1 AGUUGGUGUAGGAGGGGGUGGAGGUGCCAGUGGAGCAGCGGCAGGAGCAGGCUCCGCAGCAGGAUCUGGAUCCG------GAUCAGGUUCCGGAUCAGCAG .((((((...(((...(((......))).(((((((....((....))..))))))).((..((((((....))------))))..)))))..)))))).. ( -37.40) >DroEre_CAF1 22088 89 + 1 AGUAGGG------------GGAGUUGCCAGUGUAGCAGCGGUAGGAGCGAGCUCCGCGGCAGGAUCUGGAUCCGGAUGCGGAUCAGGUUCCACAUCAGCAG .......------------.....(((..((((....((.((.((((....)))))).)).(((.((.((((((....)))))).)).)))))))..))). ( -32.50) >consensus AGUUGGUGUAGGAGGGGGUGGAGGUGCCAGUGGAGCAGCGGCAGGAGCAGGCUCCGCAGCAGGAUCUGGAUCCG______GAUCAGGUUCCGGAUCAGCAG ...................((.....)).(((((((.((.......))..))))))).((..((((((((.((............)).)))))))).)).. (-28.49 = -29.05 + 0.56)



| Location | 20,205,647 – 20,205,742 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 84.69 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -19.76 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20205647 95 - 22224390 CUGCUGAUCCGGAACCUGAUC------CCGAUCCAGAUCCUGCUGCGGAGCCUGCUCCUGCCGCUGCUCCACUGGCACCUCCACCCCCUCCUACUCCAACU (((..((((.(((......))------).)))))))....(((((.(((((..((.......)).))))).).))))........................ ( -23.10) >DroSec_CAF1 33870 95 - 1 CUGCUGAUCCGGAACCUGAUC------CGGAUCCAGAUCCUGCUGCGGAGCCCGCUCCUGCCGCUGCUCCACUGGCACCUCCACCCCCUCCUACACCAACU (((..((((((((......))------)))))))))....(((((.(((((..((....))....))))).).))))........................ ( -28.40) >DroSim_CAF1 33527 95 - 1 CUGCUGAUCCGGAACCUGAUC------CGGAUCCAGAUCCUGCUGCGGAGCCUGCUCCUGCCGCUGCUCCACUGGCACCUCCACCCCCUCCUACACCAACU (((..((((((((......))------)))))))))....(((((.(((((..((.......)).))))).).))))........................ ( -29.20) >DroEre_CAF1 22088 89 - 1 CUGCUGAUGUGGAACCUGAUCCGCAUCCGGAUCCAGAUCCUGCCGCGGAGCUCGCUCCUACCGCUGCUACACUGGCAACUCC------------CCCUACU .((((..((((((.(..((((((....))))))..).))).((.((((((.......)).)))).)).)))..)))).....------------....... ( -27.20) >consensus CUGCUGAUCCGGAACCUGAUC______CGGAUCCAGAUCCUGCUGCGGAGCCCGCUCCUGCCGCUGCUCCACUGGCACCUCCACCCCCUCCUACACCAACU .....((((.(((.((............)).))).)))).(((((.(((((..((.......)).))))).).))))........................ (-19.76 = -20.33 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:18 2006