
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,200,350 – 20,200,505 |
| Length | 155 |
| Max. P | 0.950101 |

| Location | 20,200,350 – 20,200,470 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.75 |
| Mean single sequence MFE | -41.48 |
| Consensus MFE | -30.02 |
| Energy contribution | -32.30 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
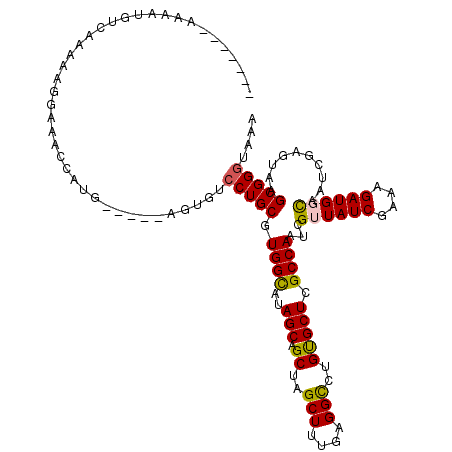
>X_DroMel_CAF1 20200350 120 - 22224390 CCUGCGUGGCAUAGCAGCUAGCUUUUAGGCCUGUGCUCGCCAAUCGUUAUCGAAAGAUGGCAAUCGAGUAAGCGGGUAAAUUGAAGUUUAGCCGCAGGCGGGUAUUAGGUUUUCAGAUUA ...((.((((......))))))....(((((((((((((((....((((((....))))))..........((((.(((((....))))).)))).)))))))).)))))))........ ( -47.00) >DroSec_CAF1 27129 120 - 1 CCUGCGUGGCAUAGCAGCUAGCUUUGAGGCCUGCGCUCGCCAAUCGUUAUCGAAAGAUGGCAAUCGAGUAAGCGGGUAAAUUGAAAUUUAGCCGCAGGCGGAUAUUAGGCUUUCAGAUUA ...((.((((......))))))(((((((((((...(((((....((((((....))))))..........((((.(((((....))))).)))).)))))....))))).))))))... ( -44.60) >DroSim_CAF1 27050 120 - 1 CCUGCGUGGCAUAGCAGCUAGCUUUGAGGCCUGCGCUCGCCAAUCGUUAUCGAAAGAUGGCAAUCGAGUAAGCGGGUAAAUUGAAAUUUAGCCGCAGGCGGACAUUAGGCUUUCAGAUUA ...((.((((......))))))(((((((((((.(..((((....((((((....))))))..........((((.(((((....))))).)))).))))..)..))))).))))))... ( -45.70) >DroEre_CAF1 17095 120 - 1 ACUGCGUGGUGUAGCAGCUAGCUUUGAGGUAUGUGCUCGCCAAUCGGUAUCGAAAGAUGGUAAUCGAGUAUGCGGCUAAAUUAAAAUGUAACAGCGGGCGGGCAUAAGAUGUUCGGGUUA .((((........)))).(((((..((.((.((((((((((..((((((((....)))....)))))...(((.((...........))....)))))))))))))..)).))..))))) ( -36.50) >DroYak_CAF1 18061 111 - 1 CCUGCGUGGCAUAGCAGCUAGCUUUCUGGCAAGUGCUCGCCAAUCGUUAUCGAAAGAUGCCAAUCGAGUAUGCGGGUAGAUUGAAAUUUAU---------GGUUUGGGGUUUCCAGAUUA ((((((((((..((((.((.(((....))).)))))).)))..(((.((((....)))).....)))..)))))))..............(---------(((((((.....)))))))) ( -33.60) >consensus CCUGCGUGGCAUAGCAGCUAGCUUUGAGGCCUGUGCUCGCCAAUCGUUAUCGAAAGAUGGCAAUCGAGUAAGCGGGUAAAUUGAAAUUUAGCCGCAGGCGGGUAUUAGGUUUUCAGAUUA ...((.((((......))))))((((((((((..(((((((....((((((....))))))..........((((.(((((....))))).)))).)))))))...)))).))))))... (-30.02 = -32.30 + 2.28)



| Location | 20,200,390 – 20,200,505 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -26.26 |
| Energy contribution | -26.14 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20200390 115 - 22224390 AGUGAAUUUAAUGUCAAAAAGGAAACCAUG-----AGUGUCCUGCGUGGCAUAGCAGCUAGCUUUUAGGCCUGUGCUCGCCAAUCGUUAUCGAAAGAUGGCAAUCGAGUAAGCGGGUAAA ..........((((((...((((.((....-----.)).))))...))))))(((.....))).....(((((((((((...((.((((((....)))))).)))))))..))))))... ( -34.90) >DroSec_CAF1 27169 115 - 1 AGUGAAUGAAAUGUCAAAAAUGAAACCAUG-----CGUGUCCUGCGUGGCAUAGCAGCUAGCUUUGAGGCCUGCGCUCGCCAAUCGUUAUCGAAAGAUGGCAAUCGAGUAAGCGGGUAAA .........................(((((-----((.....)))))))((.(((.....))).))..(((((((((((...((.((((((....)))))).)))))))..))))))... ( -34.70) >DroSim_CAF1 27090 108 - 1 -------AAAAUGUCAAAAAUGAAACCAUG-----CGUGUCCUGCGUGGCAUAGCAGCUAGCUUUGAGGCCUGCGCUCGCCAAUCGUUAUCGAAAGAUGGCAAUCGAGUAAGCGGGUAAA -------..................(((((-----((.....)))))))((.(((.....))).))..(((((((((((...((.((((((....)))))).)))))))..))))))... ( -34.70) >DroEre_CAF1 17135 111 - 1 -------AAGGUGUUAAG--GAAAACCGUGGCUGUACUGUACUGCGUGGUGUAGCAGCUAGCUUUGAGGUAUGUGCUCGCCAAUCGGUAUCGAAAGAUGGUAAUCGAGUAUGCGGCUAAA -------..(((......--....))).(((((((...(((((.(((((((.((((..((.(.....).))..)))))))))...(.((((....)))).)...)))))))))))))).. ( -32.20) >DroYak_CAF1 18092 105 - 1 -------AUAGUGUUAAA--GUA-GCCAUG-----AGCGUCCUGCGUGGCAUAGCAGCUAGCUUUCUGGCAAGUGCUCGCCAAUCGUUAUCGAAAGAUGCCAAUCGAGUAUGCGGGUAGA -------...........--...-((((.(-----(((...((((........))))...))))..))))...(((((((...(((.((((....)))).....)))....))))))).. ( -33.60) >consensus _______AAAAUGUCAAAAAGGAAACCAUG_____AGUGUCCUGCGUGGCAUAGCAGCUAGCUUUGAGGCCUGUGCUCGCCAAUCGUUAUCGAAAGAUGGCAAUCGAGUAAGCGGGUAAA ........................................(((((.((((..(((.((..(((....)))..))))).))))...((((((....))))))..........))))).... (-26.26 = -26.14 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:14 2006