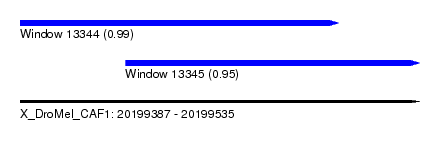
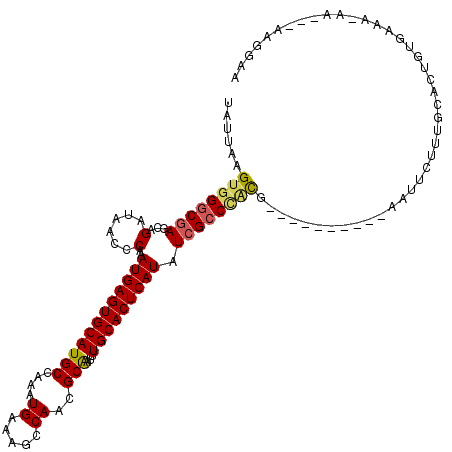
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,199,387 – 20,199,535 |
| Length | 148 |
| Max. P | 0.991534 |

| Location | 20,199,387 – 20,199,505 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.49 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -29.92 |
| Energy contribution | -30.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20199387 118 + 22224390 AAAUCAUGUGCAGCCAA-CGGCUUCGAGAGUCAUUAAGCUUAUUAAGUGGGCGAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGAAAGCCAACGCAAUUUGCACUCAUAUCGCCCACG- ........((.((((..-.)))).)).((((......)))).....((((((((...(......)..((((((((((((....((.....))..)))...))))))))).)))))))).- ( -33.70) >DroSec_CAF1 26172 118 + 1 AAAUCAUGUGCAGCCAA-CGGCUUCGAGAGUCAUUAAGUUUAUUAAGUGGGCGAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGAAAGCCAACGCGAUUUGCACUCAUAUCGCCGACG- ...((...((.((((..-.)))).)).)).................((.(((((...(......)..((((((((((((....((.....))..)))...))))))))).))))).)).- ( -27.60) >DroSim_CAF1 26096 118 + 1 AAAUCAUGUGCAGCCAA-CGGCUUCGAGAGUCAUUAAGUUUAUUAAGUGGGCGAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGAAAGCCAACGCAAUUUGCACUCAUAUCGCCCACG- ...((...((.((((..-.)))).)).)).................((((((((...(......)..((((((((((((....((.....))..)))...))))))))).)))))))).- ( -32.70) >DroEre_CAF1 16171 118 + 1 AAAUCAUGUGCAGCCAA-AGGCUUCGAGAGUCAUUAAGCUUAUUAAGUGGGCGAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGAAAGCCAACGCAAUUUGCACUCAUAUCGCCUACG- ........((.((((..-.)))).)).((((......)))).....((((((((...(......)..((((((((((((....((.....))..)))...))))))))).)))))))).- ( -31.40) >DroYak_CAF1 17154 118 + 1 AAAUCAUGUGCAGCCAA-AGGCUUCGAGAGUCAUUAAGUUUAUUAAGUGGGCGAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGGAAGCCAACGCAAUUUGCACUCAUAUCGCCCGCG- ...((...((.((((..-.)))).)).)).................((((((((...(......)..((((((((((((....(((...)))..)))...))))))))).)))))))).- ( -34.00) >DroAna_CAF1 61668 120 + 1 AAAUCUUGCUCAGCCAAAAGGCUUCGAGAGUCAUUAAGUUUAUUAAGUGGCCUAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGGAAUCCAACGCAAUUUGCACUCAUAUCGCCCCUUC ..((((.(((..((((...(((((...))))).((((.....)))).))))..))))))).......((((((((((((....(((...)))..)))...)))))))))........... ( -30.60) >consensus AAAUCAUGUGCAGCCAA_AGGCUUCGAGAGUCAUUAAGUUUAUUAAGUGGGCGAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGAAAGCCAACGCAAUUUGCACUCAUAUCGCCCACG_ ...((...((.((((....)))).)).)).................((((((((...(......)..((((((((((((....((.....))..)))...))))))))).)))))))).. (-29.92 = -30.12 + 0.20)

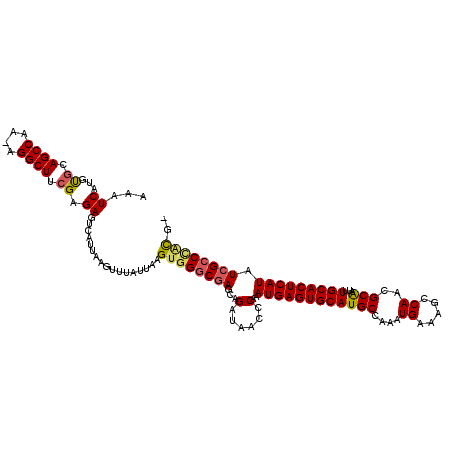
| Location | 20,199,426 – 20,199,535 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.29 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -24.46 |
| Energy contribution | -24.63 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20199426 109 + 22224390 UAUUAAGUGGGCGAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGAAAGCCAACGCAAUUUGCACUCAUAUCGCCCACG----------AAUUCCUUGCACUGUUAAA-AAAAGAAGGAA ......((((((((...(......)..((((((((((((....((.....))..)))...))))))))).)))))))).----------..((((((...........-.....)))))) ( -31.69) >DroSec_CAF1 26211 105 + 1 UAUUAAGUGGGCGAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGAAAGCCAACGCGAUUUGCACUCAUAUCGCCGACG----------AAUUCUGUGCACUGUGA-A-AA---AAGGAA ......((.(((((...(......)..((((((((((((....((.....))..)))...))))))))).))))).)).----------.................-.-..---...... ( -23.50) >DroSim_CAF1 26135 106 + 1 UAUUAAGUGGGCGAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGAAAGCCAACGCAAUUUGCACUCAUAUCGCCCACG----------AAUUCUGUGCACUGUGAGA-AA---AAGGAA ......((((((((...(......)..((((((((((((....((.....))..)))...))))))))).)))))))).----------..((((..........)))-).---...... ( -28.80) >DroEre_CAF1 16210 105 + 1 UAUUAAGUGGGCGAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGAAAGCCAACGCAAUUUGCACUCAUAUCGCCUACG----------AAUUCUUUGCACCGCGAA--AA---AACGAA ......((((((((...(......)..((((((((((((....((.....))..)))...))))))))).)))))))).----------.....(((((...)))))--..---...... ( -28.00) >DroYak_CAF1 17193 107 + 1 UAUUAAGUGGGCGAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGGAAGCCAACGCAAUUUGCACUCAUAUCGCCCGCG----------AAUUCUUUGCACCGCAAAAAAA---AACGUA ......((((((((...(......)..((((((((((((....(((...)))..)))...))))))))).)))))))).----------.....(((((...)))))....---...... ( -31.90) >DroAna_CAF1 61708 110 + 1 UAUUAAGUGGCCUAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGGAAUCCAACGCAAUUUGCACUCAUAUCGCCCCUUCAGUCCGGCCUUUUAUCUGCACUAAAAA--AA---A----- ..............((((((((.....((((((((((((....(((...)))..)))...)))))))))...(((..........)))...))))))))........--..---.----- ( -28.30) >consensus UAUUAAGUGGGCGAGCAGAUAACCCAAAUGAGUGCAUGCCAAAUGAAAGCCAACGCAAUUUGCACUCAUAUCGCCCACG__________AAUUCUUUGCACUGUGAAA_AA___AAGGAA ......((((((((...(......)..((((((((((((....((.....))..)))...))))))))).)))))))).......................................... (-24.46 = -24.63 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:11 2006