
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,191,178 – 20,191,293 |
| Length | 115 |
| Max. P | 0.912708 |

| Location | 20,191,178 – 20,191,293 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.48 |
| Mean single sequence MFE | -16.62 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.06 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20191178 115 + 22224390 --GUGUGGAAGGUGGGGAA--AAACCACAGCAAAACAAAUUAAUAAAUGCAUAAUAAAUAUGCGUAAUAAUAAUGACUAAAGAAAAAUCAGAAACAAUAAAUAUUAUUUGAGUAGUGUC --.(((.(...((((....--...))))..)...))).........(((((((.....)))))))...................................(((((((....))))))). ( -15.50) >DroSec_CAF1 12806 115 + 1 --GGUUGGAAGGUGGGGAA--AAACCACAGCAAAACAAAUUAAUAAAUGCAUAAUAAAUAUGCGUAAUAAUAAUGACUAAAGAAAAAUCAGAAACAAUAAAUAUUAUUUGAGUAGUGUC --.((((....((((....--...))))..................(((((((.....))))))).............................))))..(((((((....))))))). ( -15.90) >DroSim_CAF1 10909 114 + 1 --GGUUGGGAGGUGGGGAA---AACCACAGCAAAACAAAUUAAUAAAUGCAUAAUAAAUAUGCGUAAUAAUAAUGACUAAAGAAAAAUCAGAAACAAUAAAUAUUAUUUGAGUAGUGUC --.(((..(..((((....---..))))..)..)))..........(((((((.....)))))))...................................(((((((....))))))). ( -18.00) >DroEre_CAF1 10698 117 + 1 --AGGUGGAGAGCGGGCAAAAAAACCGCAGCAAAACAAAUUAAUAAAUGCAUAAUAAAUAUGCGUAAUAAUAAUGACUAAAGAAAAAUCAGAUACAAUAAAUAUUAUUUGAGUAGUGUC --.........((((.........)))).(((..((..........(((((((.....)))))))......................(((((((..........)))))))))..))). ( -18.50) >DroYak_CAF1 10390 117 + 1 AAGGGCGGAAAGUGGGCAA--AAACCACAGCAAAACAAAUUAAUAAAUGCAUAAUAAAUAUGCGUAAUAAUAAUGACUAAAGAAAAAUCAGAUACAAUAAAUAUUAUUUCAGUAGUGUC ...(((.....((((....--...))))..................(((((((.....)))))))..........((((..((((.....................))))..))))))) ( -15.20) >consensus __GGGUGGAAGGUGGGGAA__AAACCACAGCAAAACAAAUUAAUAAAUGCAUAAUAAAUAUGCGUAAUAAUAAUGACUAAAGAAAAAUCAGAAACAAUAAAUAUUAUUUGAGUAGUGUC ...........((((.........)))).(((..((.((.(((((.(((((((.....)))))))................((....))............))))).))..))..))). (-14.38 = -14.06 + -0.32)

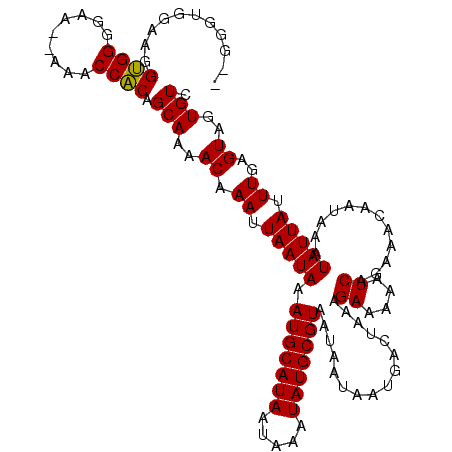
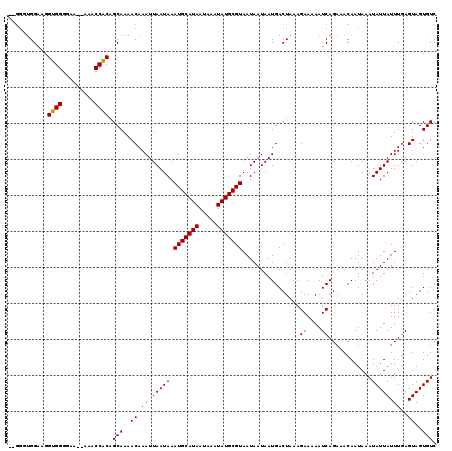
| Location | 20,191,178 – 20,191,293 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.48 |
| Mean single sequence MFE | -13.97 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.16 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20191178 115 - 22224390 GACACUACUCAAAUAAUAUUUAUUGUUUCUGAUUUUUCUUUAGUCAUUAUUAUUACGCAUAUUUAUUAUGCAUUUAUUAAUUUGUUUUGCUGUGGUUU--UUCCCCACCUUCCACAC-- ((((.......(((((((.....((...((((.......)))).)).)))))))..(((((.....)))))...........))))....(((((...--...........))))).-- ( -14.14) >DroSec_CAF1 12806 115 - 1 GACACUACUCAAAUAAUAUUUAUUGUUUCUGAUUUUUCUUUAGUCAUUAUUAUUACGCAUAUUUAUUAUGCAUUUAUUAAUUUGUUUUGCUGUGGUUU--UUCCCCACCUUCCAACC-- ...(((((.(((((((((..((.((...((((.......)))).)).)).))))).(((((.....)))))..............))))..)))))..--.................-- ( -13.60) >DroSim_CAF1 10909 114 - 1 GACACUACUCAAAUAAUAUUUAUUGUUUCUGAUUUUUCUUUAGUCAUUAUUAUUACGCAUAUUUAUUAUGCAUUUAUUAAUUUGUUUUGCUGUGGUU---UUCCCCACCUCCCAACC-- ((((.......(((((((.....((...((((.......)))).)).)))))))..(((((.....)))))...........)))).....((((..---....)))).........-- ( -13.70) >DroEre_CAF1 10698 117 - 1 GACACUACUCAAAUAAUAUUUAUUGUAUCUGAUUUUUCUUUAGUCAUUAUUAUUACGCAUAUUUAUUAUGCAUUUAUUAAUUUGUUUUGCUGCGGUUUUUUUGCCCGCUCUCCACCU-- ((((.......(((((((.....((...((((.......)))).)).)))))))..(((((.....)))))...........)))).....((((.........)))).........-- ( -14.60) >DroYak_CAF1 10390 117 - 1 GACACUACUGAAAUAAUAUUUAUUGUAUCUGAUUUUUCUUUAGUCAUUAUUAUUACGCAUAUUUAUUAUGCAUUUAUUAAUUUGUUUUGCUGUGGUUU--UUGCCCACUUUCCGCCCUU ...(((((.(((((((.............(((((.......)))))..........(((((.....)))))..........)))))))...)))))..--................... ( -13.80) >consensus GACACUACUCAAAUAAUAUUUAUUGUUUCUGAUUUUUCUUUAGUCAUUAUUAUUACGCAUAUUUAUUAUGCAUUUAUUAAUUUGUUUUGCUGUGGUUU__UUCCCCACCUUCCACCC__ ((((.......(((((((.....((...((((.......)))).)).)))))))..(((((.....)))))...........)))).....((((.........))))........... (-13.48 = -13.16 + -0.32)

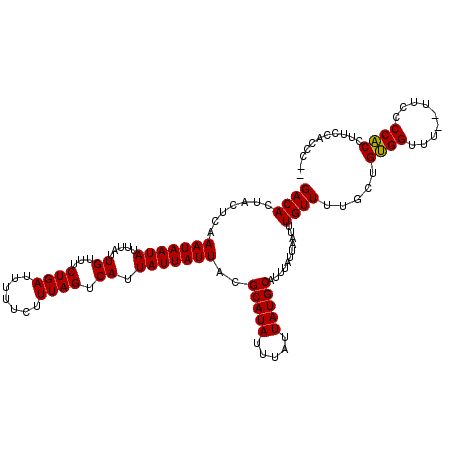
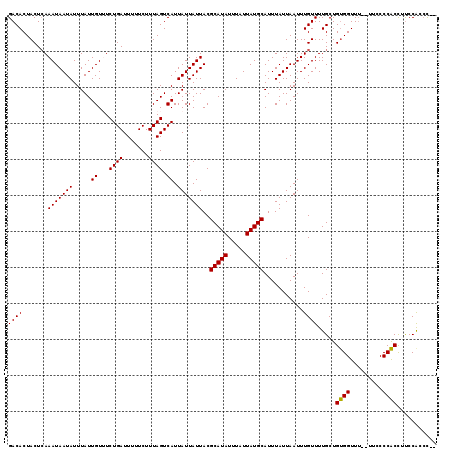
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:09 2006