
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,190,849 – 20,191,009 |
| Length | 160 |
| Max. P | 0.997509 |

| Location | 20,190,849 – 20,190,969 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -28.88 |
| Energy contribution | -29.88 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20190849 120 + 22224390 UUUGGCGGCUUUGUCGGCCUGUCGACGCACGCGGCACUAUUAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGUUUGCACAUAACUUUCAAUGAAAAGUUAAUAA ......((((.....))))(((.(((((((((((((((((((((((.(((......)))))))))))...)).)))))....)))))))).)))..(((((((......))))))).... ( -33.80) >DroSec_CAF1 12471 120 + 1 UUUGGCGGCUUUGUCGGCCUGUCGACGCACGCGGCACUAUUAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGUUUGCACAUAACUUUCAAUGAAAAGUUAAUAA ......((((.....))))(((.(((((((((((((((((((((((.(((......)))))))))))...)).)))))....)))))))).)))..(((((((......))))))).... ( -33.80) >DroSim_CAF1 10590 120 + 1 UUUGGCGGCUUUGUCGGCCUGUCGACGCACGCGGCACUAUUAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGUUUGCACAUAACUUUCAAUGAAAAGUUAAUAA ......((((.....))))(((.(((((((((((((((((((((((.(((......)))))))))))...)).)))))....)))))))).)))..(((((((......))))))).... ( -33.80) >DroEre_CAF1 10343 120 + 1 UUUGGCGGCUUUGUCGGCCUGUCGAAGCACGCGGCACUAUUAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGUUUGCACAUAACUUUCAAUGAAAAGUUAAUAA ((((((((((.....)))).))))))((((((((((((((((((((.(((......)))))))))))...)).)))))....))))).........(((((((......))))))).... ( -31.80) >DroYak_CAF1 10030 120 + 1 UAUGGCGGCUUUGUCGGCCUGUCGACGCACGCGGCACUAUUAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGUUUGCACAUAACUUUCAAUGAAAAGUUAAUAA ......((((.....))))(((.(((((((((((((((((((((((.(((......)))))))))))...)).)))))....)))))))).)))..(((((((......))))))).... ( -33.80) >DroAna_CAF1 53971 112 + 1 G--------UUUGUCGGCCUGUCGACGCACGCGGAACUAUUAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGCUGGCACAUAACUUUCAAUGAAAAGUUAAUAA .--------..(((((((((((.((((.((((......((((((((.(((......)))))))))))....).))))))).))).).)))))))..(((((((......))))))).... ( -27.80) >consensus UUUGGCGGCUUUGUCGGCCUGUCGACGCACGCGGCACUAUUAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGUUUGCACAUAACUUUCAAUGAAAAGUUAAUAA ......((((.....))))(((.(((((((((((((((((((((((.(((......)))))))))))...)).)))))....)))))))).)))..(((((((......))))))).... (-28.88 = -29.88 + 1.00)

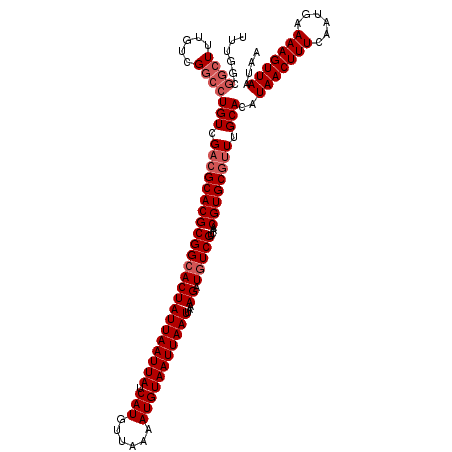

| Location | 20,190,849 – 20,190,969 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -27.41 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20190849 120 - 22224390 UUAUUAACUUUUCAUUGAAAGUUAUGUGCAAACGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUAAUAGUGCCGCGUGCGUCGACAGGCCGACAAAGCCGCCAAA ....((((((((....)))))))).((((....))))(((((((.(((.....(((((((((((......))).))))))))........)))))))))).(((.(......).)))... ( -28.82) >DroSec_CAF1 12471 120 - 1 UUAUUAACUUUUCAUUGAAAGUUAUGUGCAAACGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUAAUAGUGCCGCGUGCGUCGACAGGCCGACAAAGCCGCCAAA ....((((((((....)))))))).((((....))))(((((((.(((.....(((((((((((......))).))))))))........)))))))))).(((.(......).)))... ( -28.82) >DroSim_CAF1 10590 120 - 1 UUAUUAACUUUUCAUUGAAAGUUAUGUGCAAACGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUAAUAGUGCCGCGUGCGUCGACAGGCCGACAAAGCCGCCAAA ....((((((((....)))))))).((((....))))(((((((.(((.....(((((((((((......))).))))))))........)))))))))).(((.(......).)))... ( -28.82) >DroEre_CAF1 10343 120 - 1 UUAUUAACUUUUCAUUGAAAGUUAUGUGCAAACGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUAAUAGUGCCGCGUGCUUCGACAGGCCGACAAAGCCGCCAAA ....((((((((....))))))))((.((....((((((.((((....)....(((((((((((......))).)))))))).)).).)))))).......(((.......))))))).. ( -24.40) >DroYak_CAF1 10030 120 - 1 UUAUUAACUUUUCAUUGAAAGUUAUGUGCAAACGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUAAUAGUGCCGCGUGCGUCGACAGGCCGACAAAGCCGCCAUA ....((((((((....)))))))).((((....))))(((((((.(((.....(((((((((((......))).))))))))........)))))))))).(((.(......).)))... ( -28.82) >DroAna_CAF1 53971 112 - 1 UUAUUAACUUUUCAUUGAAAGUUAUGUGCCAGCGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUAAUAGUUCCGCGUGCGUCGACAGGCCGACAAA--------C ....((((((((....))))))))((((((.((((((((.((((....)....(((((((((((......))).)))))))).)))..)))))))).....)))..)))..--------. ( -24.80) >consensus UUAUUAACUUUUCAUUGAAAGUUAUGUGCAAACGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUAAUAGUGCCGCGUGCGUCGACAGGCCGACAAAGCCGCCAAA ....((((((((....)))))))).((((....))))(((((((.(((.....(((((((((((......))).))))))))........)))))))))).(((.......)))...... (-25.20 = -25.87 + 0.67)

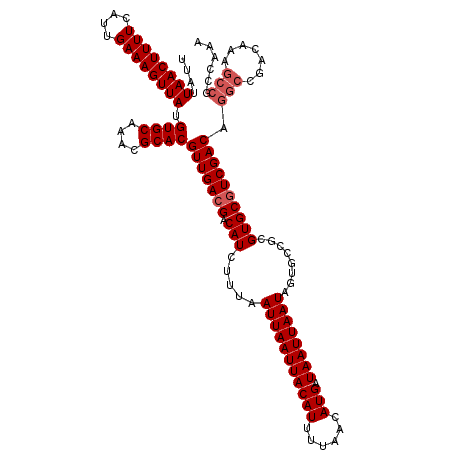

| Location | 20,190,889 – 20,191,009 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.39 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -19.81 |
| Energy contribution | -19.67 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20190889 120 + 22224390 UAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGUUUGCACAUAACUUUCAAUGAAAAGUUAAUAAAUUACGUAGUGCCGAAAACGAAAUUACAAACGCCAUCAUC ((((((.(((......)))))))))......((((.(((....((((....)))).(((((((......))))))).........(((((..((....))..)))))..))).))))... ( -23.50) >DroSec_CAF1 12511 120 + 1 UAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGUUUGCACAUAACUUUCAAUGAAAAGUUAAUAAAUUACAUAGUGCCGAAAACGAAAUUACAAACGCCAUCAUC ((((((.(((......)))))))))......((((.((....)).(((((((....(((((((......)))))))..........((((..((....))..)))))))))))))))... ( -20.00) >DroSim_CAF1 10630 120 + 1 UAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGUUUGCACAUAACUUUCAAUGAAAAGUUAAUAAAUUACAUAGUGCCGAAAACGAAAUUACAAACGCCAUCAUC ((((((.(((......)))))))))......((((.((....)).(((((((....(((((((......)))))))..........((((..((....))..)))))))))))))))... ( -20.00) >DroEre_CAF1 10383 120 + 1 UAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGUUUGCACAUAACUUUCAAUGAAAAGUUAAUAAAUUACAUAGUGCCGAAAACGAAAUUACAAACGCCAUCAAC ((((((.(((......)))))))))......((((.((....)).(((((((....(((((((......)))))))..........((((..((....))..)))))))))))))))... ( -20.00) >DroYak_CAF1 10070 120 + 1 UAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGUUUGCACAUAACUUUCAAUGAAAAGUUAAUAAAUUACAUAGUGCCGAAAACGAAAUUACAAACGCCAUCAAC ((((((.(((......)))))))))......((((.((....)).(((((((....(((((((......)))))))..........((((..((....))..)))))))))))))))... ( -20.00) >DroAna_CAF1 54003 120 + 1 UAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGCUGGCACAUAACUUUCAAUGAAAAGUUAAUAAAUUACAUAAUGCCGAAAACGAAAUUACAAACGCCAUCAAC ((((((.(((......)))))))))......((((.(((....((((....)))).(((((((......)))))))..........((((..((....))..))))...))).))))... ( -20.10) >consensus UAAUUAUCAUGUUAAAAUGUAAUUAAUUAAAGAUGUCGUCAACGUGCGUUUGCACAUAACUUUCAAUGAAAAGUUAAUAAAUUACAUAGUGCCGAAAACGAAAUUACAAACGCCAUCAAC ((((((.(((......)))))))))......((((.(((....((((....)))).(((((((......)))))))..........((((..((....))..))))...))).))))... (-19.81 = -19.67 + -0.14)

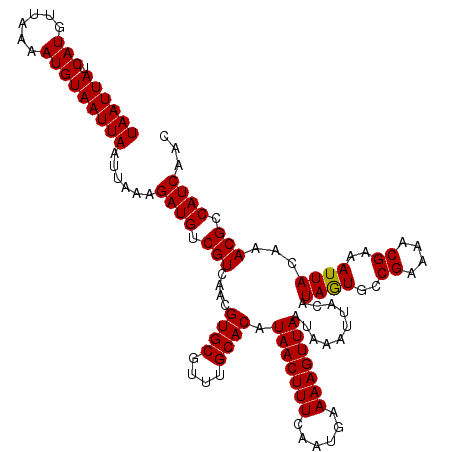
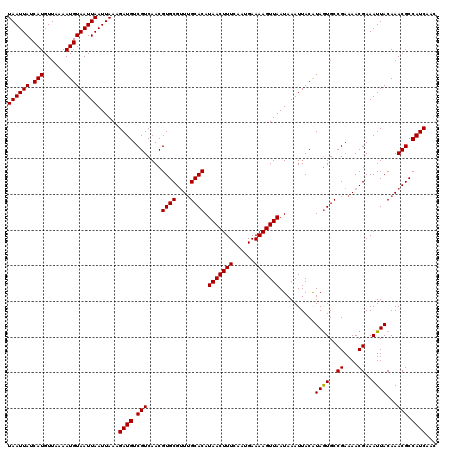
| Location | 20,190,889 – 20,191,009 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.39 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -22.57 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20190889 120 - 22224390 GAUGAUGGCGUUUGUAAUUUCGUUUUCGGCACUACGUAAUUUAUUAACUUUUCAUUGAAAGUUAUGUGCAAACGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUA ...(((((((((((((..........((......))........((((((((....))))))))..))))))))).((....)).))))......(((((((((......))).)))))) ( -23.20) >DroSec_CAF1 12511 120 - 1 GAUGAUGGCGUUUGUAAUUUCGUUUUCGGCACUAUGUAAUUUAUUAACUUUUCAUUGAAAGUUAUGUGCAAACGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUA ...(((((((((((((...(((....)))...............((((((((....))))))))..))))))))).((....)).))))......(((((((((......))).)))))) ( -23.10) >DroSim_CAF1 10630 120 - 1 GAUGAUGGCGUUUGUAAUUUCGUUUUCGGCACUAUGUAAUUUAUUAACUUUUCAUUGAAAGUUAUGUGCAAACGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUA ...(((((((((((((...(((....)))...............((((((((....))))))))..))))))))).((....)).))))......(((((((((......))).)))))) ( -23.10) >DroEre_CAF1 10383 120 - 1 GUUGAUGGCGUUUGUAAUUUCGUUUUCGGCACUAUGUAAUUUAUUAACUUUUCAUUGAAAGUUAUGUGCAAACGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUA ((..((((((((((((...(((....)))...............((((((((....))))))))..))))))))).)))..))............(((((((((......))).)))))) ( -26.20) >DroYak_CAF1 10070 120 - 1 GUUGAUGGCGUUUGUAAUUUCGUUUUCGGCACUAUGUAAUUUAUUAACUUUUCAUUGAAAGUUAUGUGCAAACGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUA ((..((((((((((((...(((....)))...............((((((((....))))))))..))))))))).)))..))............(((((((((......))).)))))) ( -26.20) >DroAna_CAF1 54003 120 - 1 GUUGAUGGCGUUUGUAAUUUCGUUUUCGGCAUUAUGUAAUUUAUUAACUUUUCAUUGAAAGUUAUGUGCCAGCGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUA ((..((((((((.........(....)(((((............((((((((....)))))))).)))))))))).)))..))............(((((((((......))).)))))) ( -23.12) >consensus GAUGAUGGCGUUUGUAAUUUCGUUUUCGGCACUAUGUAAUUUAUUAACUUUUCAUUGAAAGUUAUGUGCAAACGCACGUUGACGACAUCUUUAAUUAAUUACAUUUUAACAUGAUAAUUA ...(((((((((((((...(((....)))...............((((((((....))))))))..))))))))).((....)).))))......(((((((((......))).)))))) (-22.57 = -22.60 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:07 2006