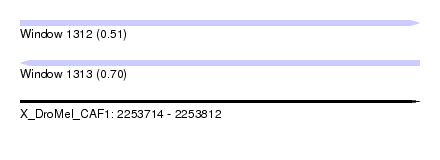
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,253,714 – 2,253,812 |
| Length | 98 |
| Max. P | 0.704252 |

| Location | 2,253,714 – 2,253,812 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -23.82 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2253714 98 + 22224390 CAAUGUGCCACCAGCAACAGUGCAACGUUGCA--------ACAUCGCAACAUCGCAGAUGCACUGCAGCCAUU-ACUCAGCCUUACAUGGACAU------GGGGUGGCUAGCU ......((((((.((..((((((...(((((.--------.....)))))(((...)))))))))..))....-..(((.((......))...)------))))))))..... ( -32.60) >DroSec_CAF1 16544 98 + 1 CAAUGUGCCACCAGCAACAGUGCAACAUUGCA--------ACAUCGCAACAUCGCAGAUGCACUGCAGCCAUU-ACUCAGCCUUACAUGGACAU------GGGGUGGCUAGCU ......((((((.((..(((((((...((((.--------.............)))).)))))))..))....-..(((.((......))...)------))))))))..... ( -29.24) >DroYak_CAF1 17601 113 + 1 CAAUGUGCCACCAGCAACAGUGCAACAUUUCAGCCAUUCCACAUUGCAACAUCGCAGAUGCAGUGCAGCCAUUGACUCGGCCUUACAUGGACAUGGAAAUGGGGUGGCUGGCU ......((((((.(((....)))..(((((...(((((((((((((((.(......).)))))))..(((........)))......)))).))))))))).))))))..... ( -36.10) >consensus CAAUGUGCCACCAGCAACAGUGCAACAUUGCA________ACAUCGCAACAUCGCAGAUGCACUGCAGCCAUU_ACUCAGCCUUACAUGGACAU______GGGGUGGCUAGCU ......((((((.((..(((((((...((((......................)))).)))))))..))..........(((......)).)..........))))))..... (-23.82 = -24.48 + 0.67)

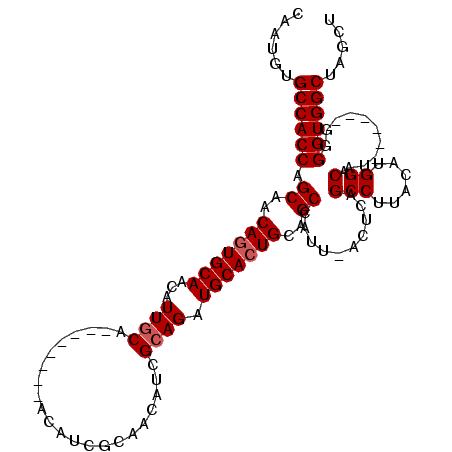
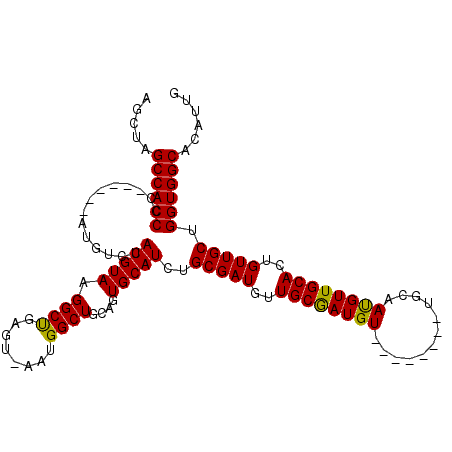
| Location | 2,253,714 – 2,253,812 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -32.23 |
| Energy contribution | -31.57 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2253714 98 - 22224390 AGCUAGCCACCCC------AUGUCCAUGUAAGGCUGAGU-AAUGGCUGCAGUGCAUCUGCGAUGUUGCGAUGU--------UGCAACGUUGCACUGUUGCUGGUGGCACAUUG .....((((((.(------(.(((.......)))))...-...(((.((((((((......((((((((....--------)))))))))))))))).)))))))))...... ( -36.70) >DroSec_CAF1 16544 98 - 1 AGCUAGCCACCCC------AUGUCCAUGUAAGGCUGAGU-AAUGGCUGCAGUGCAUCUGCGAUGUUGCGAUGU--------UGCAAUGUUGCACUGUUGCUGGUGGCACAUUG .....((((((.(------(.(((.......)))))...-...(((.((((((((..((((((((....))))--------))))....)))))))).)))))))))...... ( -36.50) >DroYak_CAF1 17601 113 - 1 AGCCAGCCACCCCAUUUCCAUGUCCAUGUAAGGCCGAGUCAAUGGCUGCACUGCAUCUGCGAUGUUGCAAUGUGGAAUGGCUGAAAUGUUGCACUGUUGCUGGUGGCACAUUG .....((((((.(((((((((.(((((((..(((((......)))))(((..(((((...)))))))).)))))))))))...)))))..(((....))).))))))...... ( -36.60) >consensus AGCUAGCCACCCC______AUGUCCAUGUAAGGCUGAGU_AAUGGCUGCAGUGCAUCUGCGAUGUUGCGAUGU________UGCAAUGUUGCACUGUUGCUGGUGGCACAUUG .....((((((..............(((((.((((........))))....)))))..(((((..((((((((............))))))))..))))).))))))...... (-32.23 = -31.57 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:22 2006