
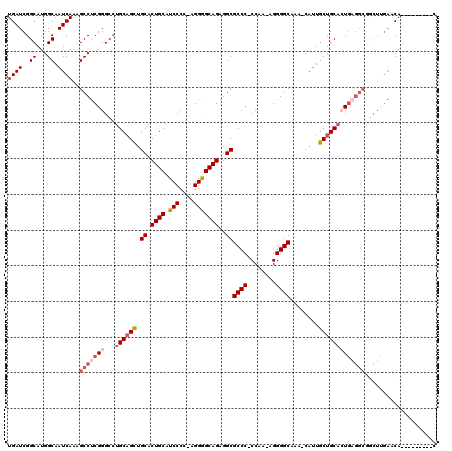
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,181,534 – 20,181,714 |
| Length | 180 |
| Max. P | 0.999889 |

| Location | 20,181,534 – 20,181,643 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.46 |
| Mean single sequence MFE | -46.42 |
| Consensus MFE | -32.80 |
| Energy contribution | -34.40 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20181534 109 - 22224390 UGAUCGGCAUGGCAAUCAAAGCCUCGGGGCUGCAGCUGCACUGCAUCCCC-AGGGGCAGAGGCGCCCCCCAA-AGGGGCAAACCAGUGCUGCACUGCGGCGUCUUGAACA---------C ((((..((...)).))))..(((((((((.(((((.....))))).))))-.))))).((((((((((....-.)))))........(((((...)))))))))).....---------. ( -48.30) >DroSec_CAF1 3161 116 - 1 UGAUCGGCAUGGCAAUCAAAGCCUCGGGCCUGCAGCUGCACUGCAUCCCC-AGGGGCAGAGGCGCCC-CCAA-AGGGGCAAG-CAUUGCUGCACUGAGGCGGCUUGAACAGAUGGUUAAC ((((((...((....((((.(((((((...(((((((((.((((..((..-..))))))....((((-(...-.)))))..)-))..)))))))))))))...)))).))..)))))).. ( -49.00) >DroSim_CAF1 2607 116 - 1 UGAUCGGCAUGGCAAUCAAAGCCUCGGGCCUGCAGCUGCACUGCAUCCCC-AGGGGCAGAGGCGCCC-CCAA-AGGGGCAAG-CAUUGCUGCACUGAGGCGGCUUGAACAGAUGGUUAAC ((((((...((....((((.(((((((...(((((((((.((((..((..-..))))))....((((-(...-.)))))..)-))..)))))))))))))...)))).))..)))))).. ( -49.00) >DroEre_CAF1 1566 107 - 1 UGAUCGGCAUGGCAAUCAAAGCCUCGGGCAUGCUGUUGCACUGCAUCCCC-AGGAGCAGAGGCGCCC-CCAA-AGGGGCGAA-CAUUGCUGCAGUGAGGCCGCUGGAGCA---------C ...(((((..(((.......)))...(((..(((((.(((((((.(((..-.)))))))...(((((-(...-.))))))..-...))).)))))...))))))))....---------. ( -45.50) >DroYak_CAF1 3128 97 - 1 UGAUCGGCAUGGCAAUCAAAGCCCCGGGCCAGCAGCUGCACUGCCUCCCCAAGGAGCAGAGGCGCCC-CCAAAAGGGGCAAA-CAUUGCUGCAGU------------GCA---------C .....(((..(((.......)))....))).(((((.((.((((.(((....)))))))..))((((-(.....)))))...-....)))))...------------...---------. ( -40.30) >consensus UGAUCGGCAUGGCAAUCAAAGCCUCGGGCCUGCAGCUGCACUGCAUCCCC_AGGGGCAGAGGCGCCC_CCAA_AGGGGCAAA_CAUUGCUGCACUGAGGCGGCUUGAACA_________C ((((..((...)).))))..(((((((...((((((.((.((((.(((....)))))))..))((((........))))........))))))))))))).................... (-32.80 = -34.40 + 1.60)


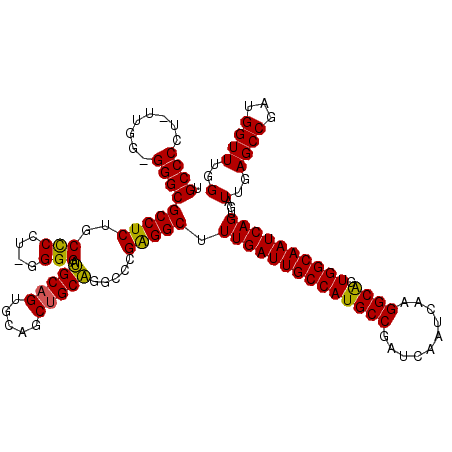
| Location | 20,181,565 – 20,181,683 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -56.38 |
| Consensus MFE | -50.92 |
| Energy contribution | -50.24 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20181565 118 + 22224390 UGCCCCU-UUGGGGGGCGCCUCUGCCCCU-GGGGAUGCAGUGCAGCUGCAGCCCCGAGGCUUUGAUUGCCAUGCCGAUCAAUCAAGGCAGUGGCAAUCAGUGAUUGAGCCGAUGGUUUGG .(((((.-....)))))(((((.......-((((.(((((.....))))).))))))))).((((((((((((((..........)))).))))))))))...(..((((...))))..) ( -57.91) >DroSec_CAF1 3200 117 + 1 UGCCCCU-UUGG-GGGCGCCUCUGCCCCU-GGGGAUGCAGUGCAGCUGCAGGCCCGAGGCUUUGAUUGCCAUGCCGAUCAAUCAAGGCGGUGGCAAUCAGUGAUUGAGCCGAUGGUUUGG .(((((.-...)-))))(((((.((.(((-.(((.(((((.....)))))..))).)))..((((((((((((((..........))).))))))))))).......)).)).))).... ( -55.50) >DroSim_CAF1 2646 117 + 1 UGCCCCU-UUGG-GGGCGCCUCUGCCCCU-GGGGAUGCAGUGCAGCUGCAGGCCCGAGGCUUUGAUUGCCAUGCCGAUCAAUCAAGGCAGUGGCAAUCAGUGAUUGAGCCGAUGGUUUGG .(((((.-...)-))))(((((.((.(((-.(((.(((((.....)))))..))).)))..((((((((((((((..........)))).)))))))))).......)).)).))).... ( -55.90) >DroEre_CAF1 1596 117 + 1 CGCCCCU-UUGG-GGGCGCCUCUGCUCCU-GGGGAUGCAGUGCAACAGCAUGCCCGAGGCUUUGAUUGCCAUGCCGAUCAAUCAAGGCAGUGGCAAUCAGUGAUUGAGCCGACGGUUUGG .(((((.-...)-))))(((((.((((((-.(((((((.(.....).)))).))).))...((((((((((((((..........)))).)))))))))).....)))).)).))).... ( -53.40) >DroYak_CAF1 3146 119 + 1 UGCCCCUUUUGG-GGGCGCCUCUGCUCCUUGGGGAGGCAGUGCAGCUGCUGGCCCGGGGCUUUGAUUGCCAUGCCGAUCAAUCAAGGCAGUGGCAAUCAGUGAUUGAGCCGAUGGUUUGG .(((((.....)-))))(((((.((((((((((..(((((.....)))))..))))))...((((((((((((((..........)))).)))))))))).....)))).)).))).... ( -59.20) >consensus UGCCCCU_UUGG_GGGCGCCUCUGCCCCU_GGGGAUGCAGUGCAGCUGCAGGCCCGAGGCUUUGAUUGCCAUGCCGAUCAAUCAAGGCAGUGGCAAUCAGUGAUUGAGCCGAUGGUUUGG .((((........))))(((((..(((....))).(((((.....))))).....))))).((((((((((((((..........)))).))))))))))...(..((((...))))..) (-50.92 = -50.24 + -0.68)


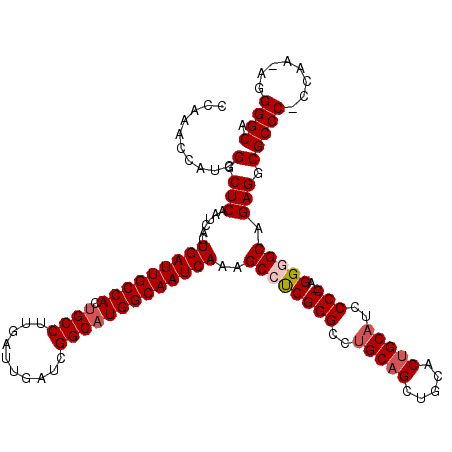
| Location | 20,181,565 – 20,181,683 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -54.24 |
| Consensus MFE | -46.38 |
| Energy contribution | -47.22 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.51 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20181565 118 - 22224390 CCAAACCAUCGGCUCAAUCACUGAUUGCCACUGCCUUGAUUGAUCGGCAUGGCAAUCAAAGCCUCGGGGCUGCAGCUGCACUGCAUCCCC-AGGGGCAGAGGCGCCCCCCAA-AGGGGCA ..........(.(((......(((((((((.((((..........)))))))))))))..(((((((((.(((((.....))))).))))-.))))).))).)(((((....-.))))). ( -60.20) >DroSec_CAF1 3200 117 - 1 CCAAACCAUCGGCUCAAUCACUGAUUGCCACCGCCUUGAUUGAUCGGCAUGGCAAUCAAAGCCUCGGGCCUGCAGCUGCACUGCAUCCCC-AGGGGCAGAGGCGCCC-CCAA-AGGGGCA ..........(.(((......(((((((((..(((..........))).)))))))))..((((((((..(((((.....)))))..)))-.))))).))).)((((-(...-.))))). ( -54.50) >DroSim_CAF1 2646 117 - 1 CCAAACCAUCGGCUCAAUCACUGAUUGCCACUGCCUUGAUUGAUCGGCAUGGCAAUCAAAGCCUCGGGCCUGCAGCUGCACUGCAUCCCC-AGGGGCAGAGGCGCCC-CCAA-AGGGGCA ..........(.(((......(((((((((.((((..........)))))))))))))..((((((((..(((((.....)))))..)))-.))))).))).)((((-(...-.))))). ( -56.40) >DroEre_CAF1 1596 117 - 1 CCAAACCGUCGGCUCAAUCACUGAUUGCCACUGCCUUGAUUGAUCGGCAUGGCAAUCAAAGCCUCGGGCAUGCUGUUGCACUGCAUCCCC-AGGAGCAGAGGCGCCC-CCAA-AGGGGCG .......(((.((((......(((((((((.((((..........)))))))))))))....((.(((.((((.(.....).))))))).-))))))...)))((((-(...-.))))). ( -50.30) >DroYak_CAF1 3146 119 - 1 CCAAACCAUCGGCUCAAUCACUGAUUGCCACUGCCUUGAUUGAUCGGCAUGGCAAUCAAAGCCCCGGGCCAGCAGCUGCACUGCCUCCCCAAGGAGCAGAGGCGCCC-CCAAAAGGGGCA .....((...((((.......(((((((((.((((..........))))))))))))).))))..))(((.((....)).((((.(((....))))))).)))((((-(.....))))). ( -49.80) >consensus CCAAACCAUCGGCUCAAUCACUGAUUGCCACUGCCUUGAUUGAUCGGCAUGGCAAUCAAAGCCUCGGGCCUGCAGCUGCACUGCAUCCCC_AGGGGCAGAGGCGCCC_CCAA_AGGGGCA ..........(.(((......(((((((((.((((..........)))))))))))))..((((((((..(((((.....)))))..)))..))))).))).)((((........)))). (-46.38 = -47.22 + 0.84)


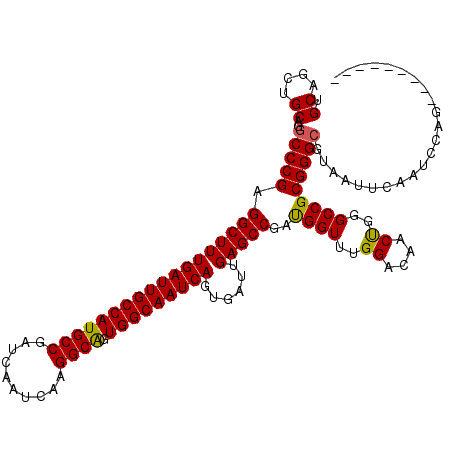
| Location | 20,181,603 – 20,181,714 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -46.32 |
| Consensus MFE | -43.40 |
| Energy contribution | -43.12 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.19 |
| SVM RNA-class probability | 0.999830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20181603 111 + 22224390 UGCAGCUGCAGCCCCGAGGCUUUGAUUGCCAUGCCGAUCAAUCAAGGCAGUGGCAAUCAGUGAUUGAGCCGAUGGUUUGGACAACUGGGCCGCGGGCGUAAUUCAAUCCAG--------- (((....)))(((((..((((((((((((((((((..........)))).)))))))))......)))))...((((..(....)..))))).))))..............--------- ( -43.60) >DroSec_CAF1 3237 111 + 1 UGCAGCUGCAGGCCCGAGGCUUUGAUUGCCAUGCCGAUCAAUCAAGGCGGUGGCAAUCAGUGAUUGAGCCGAUGGUUUGGACAACUGGGCCGCGGGCGUAAUUCAAUCCAG--------- (((....))).(((((.((((((((((((((((((..........))).)))))))))))((.(..((((...))))..).))....)))).)))))..............--------- ( -45.20) >DroSim_CAF1 2683 111 + 1 UGCAGCUGCAGGCCCGAGGCUUUGAUUGCCAUGCCGAUCAAUCAAGGCAGUGGCAAUCAGUGAUUGAGCCGAUGGUUUGGACAACCGGGCCGCGGGCGUAAUUCAAUCCAG--------- (((....))).(((((.((((((((((((((((((..........)))).)))))))))......)))))...(((((((....))))))).)))))..............--------- ( -47.30) >DroEre_CAF1 1633 120 + 1 UGCAACAGCAUGCCCGAGGCUUUGAUUGCCAUGCCGAUCAAUCAAGGCAGUGGCAAUCAGUGAUUGAGCCGACGGUUUGGACAACUGGGCCGCGGGCGUAAUUCAAUCCGGCUCCAGCUC .((....))(((((((.((((((((((((((((((..........)))).)))))))))......)))))..(((((..(....)..))))))))))))..........(((....))). ( -49.30) >DroYak_CAF1 3185 111 + 1 UGCAGCUGCUGGCCCGGGGCUUUGAUUGCCAUGCCGAUCAAUCAAGGCAGUGGCAAUCAGUGAUUGAGCCGAUGGUUUGGACAACUGCGCCGCGGGCGUAAUUUAAUCCGG--------- .((....))..(((((.(((..(((((((((((((..........)))).)))))))))((..(((..((((....)))).)))..))))).)))))..............--------- ( -46.20) >consensus UGCAGCUGCAGGCCCGAGGCUUUGAUUGCCAUGCCGAUCAAUCAAGGCAGUGGCAAUCAGUGAUUGAGCCGAUGGUUUGGACAACUGGGCCGCGGGCGUAAUUCAAUCCAG_________ .((....))..(((((.((((((((((((((((((..........)))).)))))))))......)))))..((((..((....))..)))))))))....................... (-43.40 = -43.12 + -0.28)

| Location | 20,181,603 – 20,181,714 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -40.55 |
| Consensus MFE | -37.22 |
| Energy contribution | -37.62 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20181603 111 - 22224390 ---------CUGGAUUGAAUUACGCCCGCGGCCCAGUUGUCCAAACCAUCGGCUCAAUCACUGAUUGCCACUGCCUUGAUUGAUCGGCAUGGCAAUCAAAGCCUCGGGGCUGCAGCUGCA ---------..............((..(((((((.(((.....)))....((((.......(((((((((.((((..........))))))))))))).))))...))))))).)).... ( -39.10) >DroSec_CAF1 3237 111 - 1 ---------CUGGAUUGAAUUACGCCCGCGGCCCAGUUGUCCAAACCAUCGGCUCAAUCACUGAUUGCCACCGCCUUGAUUGAUCGGCAUGGCAAUCAAAGCCUCGGGCCUGCAGCUGCA ---------..............(((((.(((...((((.((........))..))))...(((((((((..(((..........))).)))))))))..))).)))))..((....)). ( -37.00) >DroSim_CAF1 2683 111 - 1 ---------CUGGAUUGAAUUACGCCCGCGGCCCGGUUGUCCAAACCAUCGGCUCAAUCACUGAUUGCCACUGCCUUGAUUGAUCGGCAUGGCAAUCAAAGCCUCGGGCCUGCAGCUGCA ---------..............(((((.(((..(((((.((........))..)))))..(((((((((.((((..........)))))))))))))..))).)))))..((....)). ( -42.50) >DroEre_CAF1 1633 120 - 1 GAGCUGGAGCCGGAUUGAAUUACGCCCGCGGCCCAGUUGUCCAAACCGUCGGCUCAAUCACUGAUUGCCACUGCCUUGAUUGAUCGGCAUGGCAAUCAAAGCCUCGGGCAUGCUGUUGCA ..(((.(((.(.((((((.....(((.((((..............)))).)))))))))..(((((((((.((((..........)))))))))))))..).))).))).(((....))) ( -44.44) >DroYak_CAF1 3185 111 - 1 ---------CCGGAUUAAAUUACGCCCGCGGCGCAGUUGUCCAAACCAUCGGCUCAAUCACUGAUUGCCACUGCCUUGAUUGAUCGGCAUGGCAAUCAAAGCCCCGGGCCAGCAGCUGCA ---------..............(((((.((((..((((.((........))..))))..)(((((((((.((((..........)))))))))))))..))).)))))..((....)). ( -39.70) >consensus _________CUGGAUUGAAUUACGCCCGCGGCCCAGUUGUCCAAACCAUCGGCUCAAUCACUGAUUGCCACUGCCUUGAUUGAUCGGCAUGGCAAUCAAAGCCUCGGGCCUGCAGCUGCA .......................(((((.(((...((((.((........))..))))...(((((((((.((((..........)))))))))))))..))).)))))..((....)). (-37.22 = -37.62 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:58 2006