
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,181,074 – 20,181,225 |
| Length | 151 |
| Max. P | 0.981205 |

| Location | 20,181,074 – 20,181,185 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 93.62 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -27.48 |
| Energy contribution | -27.73 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20181074 111 + 22224390 UCCUGCGGGUAAAUAUUUAAGGAUGUGUUACCACCUAUUUACCAUCGUUUCUCCUUGGCGGCUGCAAGUGCCGCCUAAGCCAUCCCAUCCCAUC----CAGCUCAGAGGAUACGA .......(((((((.....(((.((......)))))))))))).((((...(((((((((((.......))))))..(((..............----..)))..))))).)))) ( -28.79) >DroSec_CAF1 2726 115 + 1 UCCUCCGGGUAAAUAUUUAAGGAUGUGUUACCACCUAUUUACCAUCGUUUCUCCUUGGCGGCUGCAAGUGCCGCCUAAGCCAUCCCAUCCCAUCACAUCCAUUCAGAGGAUCCGA (((((.((((((((.....(((.((......))))))))))))..........(((((((((.......)))))).)))........................).)))))..... ( -28.50) >DroSim_CAF1 2152 115 + 1 UCCUCCGGGUAAAUAUUUAAGGAUGUGUUACCACCUAUUUACCAUCGUUUCUCCUUGGCGGCUGCAAGUGCCGCCUAAGCCAUCCCAUCCCAUCACAUCCACUCAGAGGAUCCGA (((((.((((..........(((((((.............((....)).....(((((((((.......)))))).)))..............))))))))))).)))))..... ( -30.80) >DroEre_CAF1 1152 115 + 1 UCCUCCGGGUAAAUAUUUAAGGAUGUGUUACCACCUAUUUACCAUCAUUUCUCCUUGGCGGCUGCAAGUGCCGCCUAAGCCAGCCCAUCCCAUUCCAACUAGUCAGAGGAUCCGA (((((.((((((((.....(((.((......))))))))))))..........(((((((((.......)))))).)))........................).)))))..... ( -28.50) >consensus UCCUCCGGGUAAAUAUUUAAGGAUGUGUUACCACCUAUUUACCAUCGUUUCUCCUUGGCGGCUGCAAGUGCCGCCUAAGCCAUCCCAUCCCAUCACAUCCACUCAGAGGAUCCGA (((((.((((((((.....(((.((......))))))))))))..........(((((((((.......)))))).)))........................).)))))..... (-27.48 = -27.73 + 0.25)



| Location | 20,181,074 – 20,181,185 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 93.62 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -36.04 |
| Energy contribution | -38.73 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20181074 111 - 22224390 UCGUAUCCUCUGAGCUG----GAUGGGAUGGGAUGGCUUAGGCGGCACUUGCAGCCGCCAAGGAGAAACGAUGGUAAAUAGGUGGUAACACAUCCUUAAAUAUUUACCCGCAGGA ((((.((((.(((((((----..(......)..)))))))((((((.......)))))).))))...)))).((((((((((((......))))......))))))))....... ( -34.00) >DroSec_CAF1 2726 115 - 1 UCGGAUCCUCUGAAUGGAUGUGAUGGGAUGGGAUGGCUUAGGCGGCACUUGCAGCCGCCAAGGAGAAACGAUGGUAAAUAGGUGGUAACACAUCCUUAAAUAUUUACCCGGAGGA .....(((((((..(((((((...((((((..((.(((((((((((.......))))))..(......).........))))).))....))))))...)))))))..))))))) ( -42.60) >DroSim_CAF1 2152 115 - 1 UCGGAUCCUCUGAGUGGAUGUGAUGGGAUGGGAUGGCUUAGGCGGCACUUGCAGCCGCCAAGGAGAAACGAUGGUAAAUAGGUGGUAACACAUCCUUAAAUAUUUACCCGGAGGA .....(((((((.((((((((...((((((..((.(((((((((((.......))))))..(......).........))))).))....))))))...)))))))).))))))) ( -46.10) >DroEre_CAF1 1152 115 - 1 UCGGAUCCUCUGACUAGUUGGAAUGGGAUGGGCUGGCUUAGGCGGCACUUGCAGCCGCCAAGGAGAAAUGAUGGUAAAUAGGUGGUAACACAUCCUUAAAUAUUUACCCGGAGGA .....(((((((.((((((.(.......).))))))(((.((((((.......)))))).))).........((((((((((((......))))......))))))))))))))) ( -36.50) >consensus UCGGAUCCUCUGAGUGGAUGUGAUGGGAUGGGAUGGCUUAGGCGGCACUUGCAGCCGCCAAGGAGAAACGAUGGUAAAUAGGUGGUAACACAUCCUUAAAUAUUUACCCGGAGGA .....(((((((.((((((((...((((((..((.(((((((((((.......))))))..(......).........))))).))....))))))...)))))))).))))))) (-36.04 = -38.73 + 2.69)



| Location | 20,181,114 – 20,181,225 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -25.96 |
| Energy contribution | -30.52 |
| Covariance contribution | 4.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
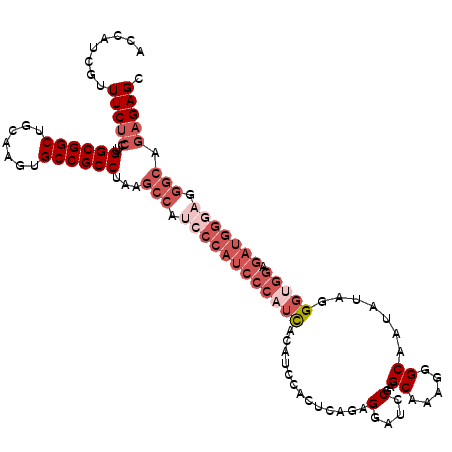
>X_DroMel_CAF1 20181114 111 + 22224390 ACCAUCGUUUCUCCUUGGCGGCUGCAAGUGCCGCCUAAGCCAUCCCAUCCCAUC----CAGCUCAGAGGAUACGAGCAAAGGGCAAUAUUGGGUGGAGAUGGGAGGGCAGAGAGC ........(((((...((((((.......))))))...(((.((((((((((((----((((((.........))))............))))))).))))))).))).))))). ( -49.90) >DroSec_CAF1 2766 96 + 1 ACCAUCGUUUCUCCUUGGCGGCUGCAAGUGCCGCCUAAGCCAUCCCAUCCCAUCACAUCCAUUCAGAGGAUCCGAGCAAAGGGCAAUAUAGG-------------------GAGU ..........((((((((((((.......))))))...(((......((..(((...((......)).)))..))......))).....)))-------------------))). ( -28.00) >DroSim_CAF1 2192 115 + 1 ACCAUCGUUUCUCCUUGGCGGCUGCAAGUGCCGCCUAAGCCAUCCCAUCCCAUCACAUCCACUCAGAGGAUCCGAGCAAAGGGCAAUAUAGGGCGGAGAUGGGAGGGCAGAGAGC ........(((((...((((((.......))))))...(((.(((((((((.((..((((.......))))..))((.....))..........)).))))))).))).))))). ( -43.60) >DroEre_CAF1 1192 115 + 1 ACCAUCAUUUCUCCUUGGCGGCUGCAAGUGCCGCCUAAGCCAGCCCAUCCCAUUCCAACUAGUCAGAGGAUCCGAGCAAAGGGCACAUUCGGGUGGAGAUGGCAGGGCAGAGAGC ........(((((...((((((.......))))))...(((...((((((((((((...........))).((((((.....))....)))))))).)))))...))).))))). ( -41.00) >consensus ACCAUCGUUUCUCCUUGGCGGCUGCAAGUGCCGCCUAAGCCAUCCCAUCCCAUCACAUCCACUCAGAGGAUCCGAGCAAAGGGCAAUAUAGGGUGGAGAUGGGAGGGCAGAGAGC ........(((((...((((((.......))))))...(((.((((((((((((.............(....)..((.....)).......))))).))))))).))).))))). (-25.96 = -30.52 + 4.56)


| Location | 20,181,114 – 20,181,225 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -42.88 |
| Consensus MFE | -26.55 |
| Energy contribution | -32.55 |
| Covariance contribution | 6.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20181114 111 - 22224390 GCUCUCUGCCCUCCCAUCUCCACCCAAUAUUGCCCUUUGCUCGUAUCCUCUGAGCUG----GAUGGGAUGGGAUGGCUUAGGCGGCACUUGCAGCCGCCAAGGAGAAACGAUGGU ..((((((((.(((((((.(((.(((............(((((.......)))))))----).)))))))))).)))...((((((.......))))))..)))))......... ( -47.30) >DroSec_CAF1 2766 96 - 1 ACUC-------------------CCUAUAUUGCCCUUUGCUCGGAUCCUCUGAAUGGAUGUGAUGGGAUGGGAUGGCUUAGGCGGCACUUGCAGCCGCCAAGGAGAAACGAUGGU .(((-------------------(........(((.((.((((.((((.......)))).))).).)).)))........((((((.......))))))..)))).......... ( -33.60) >DroSim_CAF1 2192 115 - 1 GCUCUCUGCCCUCCCAUCUCCGCCCUAUAUUGCCCUUUGCUCGGAUCCUCUGAGUGGAUGUGAUGGGAUGGGAUGGCUUAGGCGGCACUUGCAGCCGCCAAGGAGAAACGAUGGU ..((((((((.(((((((((........((..(..((..(((((.....)))))..)).)..))))))))))).)))...((((((.......))))))..)))))......... ( -50.10) >DroEre_CAF1 1192 115 - 1 GCUCUCUGCCCUGCCAUCUCCACCCGAAUGUGCCCUUUGCUCGGAUCCUCUGACUAGUUGGAAUGGGAUGGGCUGGCUUAGGCGGCACUUGCAGCCGCCAAGGAGAAAUGAUGGU ..((((((((..((((((.(((.((((....((.....))))))....((..(....)..)).)))))).))).)))...((((((.......))))))..)))))......... ( -40.50) >consensus GCUCUCUGCCCUCCCAUCUCCACCCUAUAUUGCCCUUUGCUCGGAUCCUCUGAGUGGAUGUGAUGGGAUGGGAUGGCUUAGGCGGCACUUGCAGCCGCCAAGGAGAAACGAUGGU ..((((((((.(((((((((........(((((...((((((((.....))))))))..)))))))))))))).)))...((((((.......))))))..)))))......... (-26.55 = -32.55 + 6.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:54 2006