| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,180,316 – 20,180,469 |
| Length | 153 |
| Max. P | 0.750574 |

| Location | 20,180,316 – 20,180,430 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -16.59 |
| Energy contribution | -17.63 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20180316 114 - 22224390 AUUGCCACCUGGGCGUCUCACACCUUACU------GAACGCCCAGUCCAAUCCGCACCAGCUGUCACCCGCAAAAUCUCCGCGAAAUCCCAGCUGAAAACUUCCCUACAAUGCGAUUUGU (((((...((((((((.(((........)------))))))))))............((((((.....(((.........)))......))))))................))))).... ( -29.90) >DroSec_CAF1 1984 114 - 1 AUUGCCACCUGGGCAUUUCACACCUUUCU------GAACGCCCAGUCCAAUCCCCGCCAGCUGUCACCCGCAAAAUCUCCGCGAAAUCCCAGCUGAAAACUUCCCUACAAUGCGAUUUGU (((((...((((((..((((........)------))).))))))............((((((.....(((.........)))......))))))................))))).... ( -26.10) >DroSim_CAF1 1402 114 - 1 AUUGCCACCUGGGCAUCUCACACCUUUCU------GAACGCCCAGUCCAAUCCCCGCCAGCUGUCACCCGCAAAAUCUCCGCGAAAUCCCAGCUGAAAACUUCCCUACAAUGCGAUUUGU (((((...((((((...(((........)------))..))))))............((((((.....(((.........)))......))))))................))))).... ( -25.20) >DroEre_CAF1 396 113 - 1 AUUGCCACCUG-CCAUCUCACACCUUUCG------AAGCGCGUAGUUCAAUCCCCACCAGCUGUCACCCGCAAAAUCUCCGCAAAAUCCCAGCUGAAAACUUCCCUACAAUGCGAUUUGU ...........-...............((------((.(((((.((...........((((((......((.........)).......))))))...........)).))))).)))). ( -16.57) >DroYak_CAF1 1988 119 - 1 AUUGCCACCUG-CCAGCUCACACUUUUCGGCAGCCACACGCCCAGUCCAAUCCUCACCAGCUGUCACCCGCGAAAUCUCCGCGAAAUCCCAGCUAAAAACUUCCCUACCAUGCGAUUUGU (((((...(((-((..............))))).........................(((((.....((((.......))))......))))).................))))).... ( -20.14) >consensus AUUGCCACCUGGGCAUCUCACACCUUUCU______GAACGCCCAGUCCAAUCCCCACCAGCUGUCACCCGCAAAAUCUCCGCGAAAUCCCAGCUGAAAACUUCCCUACAAUGCGAUUUGU (((((...((((((.........................))))))............((((((.....(((.........)))......))))))................))))).... (-16.59 = -17.63 + 1.04)


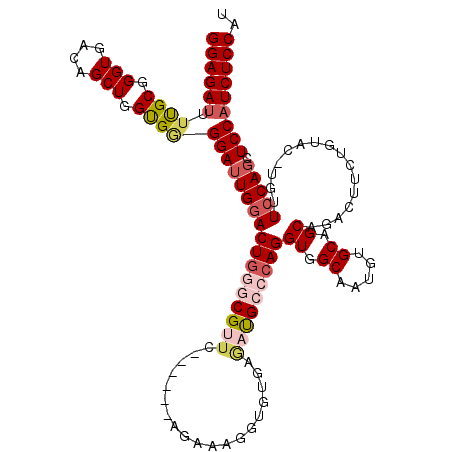
| Location | 20,180,356 – 20,180,469 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -29.91 |
| Energy contribution | -31.51 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20180356 113 + 22224390 GGAGAUUUUGCGGGUGACAGCUGGUGCGGAUUGGACUGGGCGUUC------AGUAAGGUGUGAGACGCCCAGGUGGCAAUGUGCAGCAGACUUCUGUAC-UGUUCCAGCUCCAUCUCCAU ((((((...((.((.((((((((.((((.((((..((((((((((------(........))).))))))))....)))).)))).)))((....)).)-)))))).))...)))))).. ( -49.20) >DroSec_CAF1 2024 113 + 1 GGAGAUUUUGCGGGUGACAGCUGGCGGGGAUUGGACUGGGCGUUC------AGAAAGGUGUGAAAUGCCCAGGUGGCAAUGUGCAGCAGACUUCUGUAU-UGUUCCAGCUCCAUCUCCAU ((((((...((.(((....))).))((((.(((((((((((((((------(.(....).)).)))))))))...(((((.....((((....))))))-)))))))))))))))))).. ( -46.10) >DroSim_CAF1 1442 113 + 1 GGAGAUUUUGCGGGUGACAGCUGGCGGGGAUUGGACUGGGCGUUC------AGAAAGGUGUGAGAUGCCCAGGUGGCAAUGUGCAGCAGACUUCUGUAU-UGUUCCAGCUCCAUCUCCAU ((((((...((.(((....))).))((((.(((((((((((((((------(.(....).))).))))))))...(((((.....((((....))))))-)))))))))))))))))).. ( -46.10) >DroEre_CAF1 436 113 + 1 GGAGAUUUUGCGGGUGACAGCUGGUGGGGAUUGAACUACGCGCUU------CGAAAGGUGUGAGAUGG-CAGGUGGCAAUGUGCAGCAGACUACUGUUCCAGCUCCAGCUCCUUCUCCAU (((((.......(....)((((((.(.(((.....((.((((((.------.....))))))))...(-((.((....)).))).((((....)))))))..).))))))...))))).. ( -36.80) >DroYak_CAF1 2028 108 + 1 GGAGAUUUCGCGGGUGACAGCUGGUGAGGAUUGGACUGGGCGUGUGGCUGCCGAAAAGUGUGAGCUGG-CAGGUGGCAAUGUGCAGCAGA-----------GCUCCAUCUCCAUCUCCAU ((((((.((((.(((....))).))))(((.(((((((.(((..(.((..((....(((....)))..-..))..)).)..)))..))).-----------..))))..))))))))).. ( -43.70) >consensus GGAGAUUUUGCGGGUGACAGCUGGUGGGGAUUGGACUGGGCGUUC______AGAAAGGUGUGAGAUGCCCAGGUGGCAAUGUGCAGCAGACUUCUGUAC_UGUUCCAGCUCCAUCUCCAU ((((((.((((.(((....))).))))(((((((((((((((((...................)))))))))((.((.....)).))................))))).))))))))).. (-29.91 = -31.51 + 1.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:42 2006