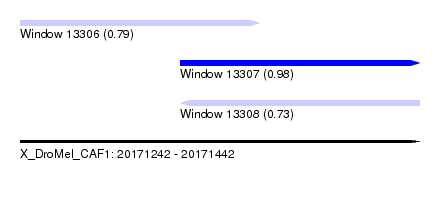
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,171,242 – 20,171,442 |
| Length | 200 |
| Max. P | 0.984968 |

| Location | 20,171,242 – 20,171,362 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.67 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -25.02 |
| Energy contribution | -24.58 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
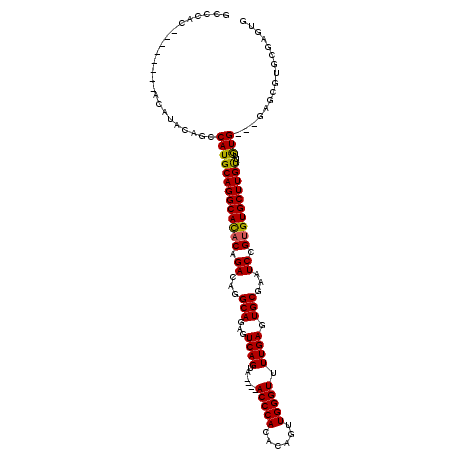
>X_DroMel_CAF1 20171242 120 + 22224390 UGCGAAAAUGUUGCCGUCAAUUCUGCACAAAAUAUGCAUAAAUACGGGCAUAAAUAAAUGUCUGGAUGGGCUCUCUCACACACUCGCACGCUCGCACAUAAACAAGCACACGGAUUCGCA .(((((..((((((((((.....((((.......))))......(((((((......))))))))))).................((......))..........))).)))..))))). ( -26.90) >DroSec_CAF1 2868 116 + 1 UGCGAAAAUGUUGCCGUCAAUUCUGCACAAAAUAUGCAUAAAUACGGGCAUAAAUAAAUGUCUGGAUGGGCUCU--CACACACUCGCACGCUCA--CACAAACAAGCACGCGGAUUCGCA .(((((.......(((((.....((((.......))))......(((((((......)))))))))))).....--.......((((..(((..--........)))..)))).))))). ( -28.40) >DroSim_CAF1 3168 100 + 1 UGCGAAUAUGUUGCCGUCAAUUCUGCACAAAAUAUGCAUAAAUACGGGCAUAAAUAAGUGUCUGGAUGGGCUCU--CA------------------CACAAGCAAGCACACGGAUUCGCA .((((((.((((((((((.....((((.......))))......(((((((......))))))))))).(((..--..------------------....)))..))).))).)))))). ( -28.40) >consensus UGCGAAAAUGUUGCCGUCAAUUCUGCACAAAAUAUGCAUAAAUACGGGCAUAAAUAAAUGUCUGGAUGGGCUCU__CACACACUCGCACGCUC___CACAAACAAGCACACGGAUUCGCA .(((((..(((..(((((.....((((.......))))......(((((((......))))))))))))(((................................)))..)))..))))). (-25.02 = -24.58 + -0.44)


| Location | 20,171,322 – 20,171,442 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -24.30 |
| Energy contribution | -24.63 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20171322 120 + 22224390 CACUCGCACGCUCGCACAUAAACAAGCACACGGAUUCGCACUCAAAACCCAACUGUGUGGGUGAGUGACUGACUCUGCAUGUCGGUAUGCCUGCAUGGCUGUAUGUAUGUGUGUAUGGGC ..((((.((((.((.(((((....(((...((((.((.(((((...(((((......))))))))))...)).))))(((((.((....)).)))))))).))))).)).)))).)))). ( -42.00) >DroSec_CAF1 2946 106 + 1 CACUCGCACGCUCA--CACAAACAAGCACGCGGAUUCGCACUCAAAACCCAACUGCGUGGGU----UACUGACUCUGCCUGUCUGUGUGCCUGCAUGGUUGUAUGU--------GUGGGC .........(((((--((((.((((((((((((((..(((.(((.((((((......)))))----)..)))...)))..))))))))))((....)))))).)))--------)))))) ( -42.90) >DroSim_CAF1 3244 92 + 1 ----------------CACAAGCAAGCACACGGAUUCGCACUCAAAACCCAACUGUGUGGGU----UACUGACUCUGCCUUUCUGUGUGCCUGCAUGGCUGUAUGU--------GUGGGC ----------------(((..(((.(((((((((...(((.(((.((((((......)))))----)..)))...)))...))))))))).)))...((.....))--------)))... ( -33.30) >consensus CACUCGCACGCUC___CACAAACAAGCACACGGAUUCGCACUCAAAACCCAACUGUGUGGGU____UACUGACUCUGCCUGUCUGUGUGCCUGCAUGGCUGUAUGU________GUGGGC ................(((......((((((((((..(((.(((..(((((......))))).......)))...)))..))))))))))..(((((....)))))........)))... (-24.30 = -24.63 + 0.34)

| Location | 20,171,322 – 20,171,442 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -24.69 |
| Energy contribution | -24.37 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20171322 120 - 22224390 GCCCAUACACACAUACAUACAGCCAUGCAGGCAUACCGACAUGCAGAGUCAGUCACUCACCCACACAGUUGGGUUUUGAGUGCGAAUCCGUGUGCUUGUUUAUGUGCGAGCGUGCGAGUG .........(((.........((.(((..((....))..))))).((.((.(.((((((((((......)))))...)))))))).))((..(((((((......)))))))..)).))) ( -35.60) >DroSec_CAF1 2946 106 - 1 GCCCAC--------ACAUACAACCAUGCAGGCACACAGACAGGCAGAGUCAGUA----ACCCACGCAGUUGGGUUUUGAGUGCGAAUCCGCGUGCUUGUUUGUG--UGAGCGUGCGAGUG ......--------........((((((...((((((((((((((.(.((((.(----(((((......)))))))))).)(((....))).))))))))))))--)).))))).).... ( -42.40) >DroSim_CAF1 3244 92 - 1 GCCCAC--------ACAUACAGCCAUGCAGGCACACAGAAAGGCAGAGUCAGUA----ACCCACACAGUUGGGUUUUGAGUGCGAAUCCGUGUGCUUGCUUGUG---------------- .....(--------(((.........((((((((((.((...(((...((((.(----(((((......)))))))))).)))...)).)))))))))).))))---------------- ( -32.20) >consensus GCCCAC________ACAUACAGCCAUGCAGGCACACAGACAGGCAGAGUCAGUA____ACCCACACAGUUGGGUUUUGAGUGCGAAUCCGUGUGCUUGUUUGUG___GAGCGUGCGAGUG .......................(((((((((((((.((...(((...((((......(((((......))))).)))).)))...)).))))))))))..)))................ (-24.69 = -24.37 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:37 2006