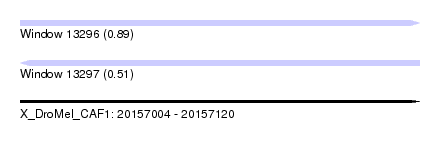
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,157,004 – 20,157,120 |
| Length | 116 |
| Max. P | 0.886579 |

| Location | 20,157,004 – 20,157,120 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -6.58 |
| Energy contribution | -5.94 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20157004 116 + 22224390 CCUUAAAGUGCAUUCGAAAUAUUACUCGUAUUUACAUGCUCAUGAAUAUUUCAUUUU-UAUCUCGAGA---CCGUCCCCACGUAAACAACUUUGGUCAAGUUGGACGUAUGUAUUUCAUU .....((((((((..((((((((....((((....)))).....)))))))).....-..........---........((((...((((((.....)))))).)))))))))))).... ( -20.70) >DroSec_CAF1 2903 110 + 1 CCUUAAAGUGCAUUCGAAAUAUUACUCGUAUUUACAUGCUCAUGAAUAUUUCAUUUU-UAUCUCGAGU---CCGUC--CAAGUAACCAACAUUGGUCAAUUUGGACGUA----UUUAAUU .....((((((....((((((((....((((....)))).....)))))))).....-..........---..(((--(((((.((((....))))..)))))))))))----))).... ( -25.10) >DroSim_CAF1 3780 110 + 1 CUUUAAAGUGCAUUCGAAAUAUUACUCGUAUUUACAUGCUCAUGAAUAUUUCAUUUU-UAUCUCGAGU---CCGUC--CAACUAACCAACAUUGGUCAAUUUGGACGUA----UUUAAUU .....((((((....((((((((....((((....)))).....)))))))).....-..........---..(((--(((...((((....))))....)))))))))----))).... ( -22.60) >DroEre_CAF1 8671 107 + 1 -------GUGCAUUUGAAAUCUUACUCGAAUUUGCAUGCCCAUGAAUGUUUAAUUUCCCAUAUCGAGUAACCAGUC--CAGGCAACCAAUUUUGGUCAAUUUAGACUCC----UUCUAUU -------.(((....((....(((((((((((.((((........))))..)))........))))))))....))--...)))((((....)))).....((((....----.)))).. ( -15.90) >DroYak_CAF1 11531 104 + 1 CCUUAAAGUGCACUCGAAAAUUCACUCGUAUCUACACGAUCGUGAAUAUUUCAUUUAUC----------UCUAGUU--CAAGUAACCAAUUUUGGUCAAUUUGCACUCA----UUUUAUU ...(((((((.....((((((((((((((......))))..)))))).)))).......----------...(((.--(((((.((((....))))..))))).)))))----))))).. ( -20.70) >consensus CCUUAAAGUGCAUUCGAAAUAUUACUCGUAUUUACAUGCUCAUGAAUAUUUCAUUUU_UAUCUCGAGU___CCGUC__CAAGUAACCAACUUUGGUCAAUUUGGACGUA____UUUAAUU .......(((.((.(((........))).)).))).....................................((((...((...((((....))))...))..))))............. ( -6.58 = -5.94 + -0.64)

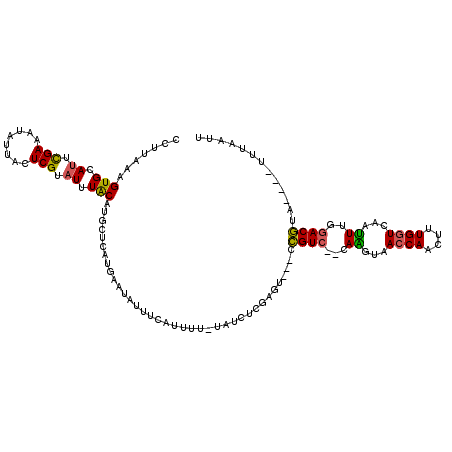
| Location | 20,157,004 – 20,157,120 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -11.76 |
| Energy contribution | -11.52 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20157004 116 - 22224390 AAUGAAAUACAUACGUCCAACUUGACCAAAGUUGUUUACGUGGGGACGG---UCUCGAGAUA-AAAAUGAAAUAUUCAUGAGCAUGUAAAUACGAGUAAUAUUUCGAAUGCACUUUAAGG ..(((((((..(((...((((((.....))))))(((((((((((....---.)))......-...((((.....))))...)))))))).....))).))))))).............. ( -20.70) >DroSec_CAF1 2903 110 - 1 AAUUAAA----UACGUCCAAAUUGACCAAUGUUGGUUACUUG--GACGG---ACUCGAGAUA-AAAAUGAAAUAUUCAUGAGCAUGUAAAUACGAGUAAUAUUUCGAAUGCACUUUAAGG ..(((((----..(((((((..((((((....)))))).)))--)))).---..((((((((-...((((.....))))..((.(((....))).))..))))))))......))))).. ( -25.30) >DroSim_CAF1 3780 110 - 1 AAUUAAA----UACGUCCAAAUUGACCAAUGUUGGUUAGUUG--GACGG---ACUCGAGAUA-AAAAUGAAAUAUUCAUGAGCAUGUAAAUACGAGUAAUAUUUCGAAUGCACUUUAAAG ..(((((----..(((((((.(((((((....))))))))))--)))).---..((((((((-...((((.....))))..((.(((....))).))..))))))))......))))).. ( -27.20) >DroEre_CAF1 8671 107 - 1 AAUAGAA----GGAGUCUAAAUUGACCAAAAUUGGUUGCCUG--GACUGGUUACUCGAUAUGGGAAAUUAAACAUUCAUGGGCAUGCAAAUUCGAGUAAGAUUUCAAAUGCAC------- .......----(((((((.....(((((....)))))(((..--....)))(((((((((((.(((........))).....)))).....))))))))))))))........------- ( -20.50) >DroYak_CAF1 11531 104 - 1 AAUAAAA----UGAGUGCAAAUUGACCAAAAUUGGUUACUUG--AACUAGA----------GAUAAAUGAAAUAUUCACGAUCGUGUAGAUACGAGUGAAUUUUCGAGUGCACUUUAAGG .......----.(((((((...((((((....))))))....--.......----------......(((((.((((((..(((((....))))))))))))))))..)))))))..... ( -25.10) >consensus AAUAAAA____UACGUCCAAAUUGACCAAAGUUGGUUACUUG__GACGG___ACUCGAGAUA_AAAAUGAAAUAUUCAUGAGCAUGUAAAUACGAGUAAUAUUUCGAAUGCACUUUAAGG .............((((..((.((((((....)))))).))...))))....................................((((....((((......))))..))))........ (-11.76 = -11.52 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:27 2006