
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,154,195 – 20,154,307 |
| Length | 112 |
| Max. P | 0.784519 |

| Location | 20,154,195 – 20,154,307 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -28.60 |
| Energy contribution | -28.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.26 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20154195 112 - 22224390 UUCCCCCCUCCCCCCCCAGUCACAGU--------UUCCAUUCCCGAUUCUCCACAUCCACUGGCGACCCAAACGCUGGAUGGCACCUAGGUGACAGUCGCUAGCAUCCGGAGGGUCCAGA .....((((((.......((...((.--------.((.......))..))..)).....(((((((((((.....))).((.(((....))).)))))))))).....))))))...... ( -30.20) >DroSec_CAF1 9084 101 - 1 UUCC-----------CCAGUCGUAGC--------UUCCAUUGCCGAUUCUCCACAUCCACUGGCGACCCAAACGCUGGAUGGCACCUAGGUGACAGUCGCUAGCAUCCGGAGGGUCCAGA ....-----------.........((--------.......)).((((((((.......(((((((((((.....))).((.(((....))).)))))))))).....)))))))).... ( -29.10) >DroYak_CAF1 7550 109 - 1 UGUC-----------CCAGUCGCAGUUUCCCAGUUUCCAUUCCCGAUUCUCCACACUCACUGGCGACCCAAACGCUGGAUGGCACCAAGGUGACAGUCGCUAGCAUCCGGAGGGUCCAGA ....-----------.............................((((((((.......(((((((((((.....))).((.(((....))).)))))))))).....)))))))).... ( -28.60) >consensus UUCC___________CCAGUCGCAGU________UUCCAUUCCCGAUUCUCCACAUCCACUGGCGACCCAAACGCUGGAUGGCACCUAGGUGACAGUCGCUAGCAUCCGGAGGGUCCAGA ............................................((((((((.......(((((((((((.....))).((.(((....))).)))))))))).....)))))))).... (-28.60 = -28.60 + -0.00)

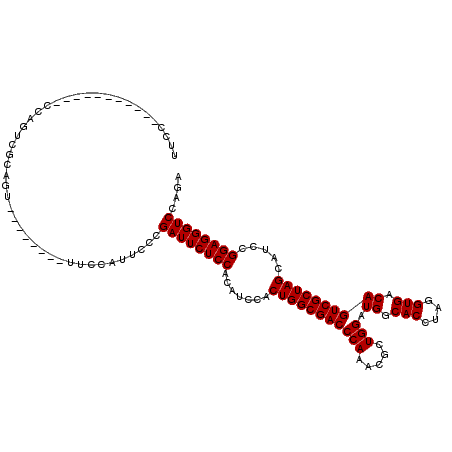
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:26 2006