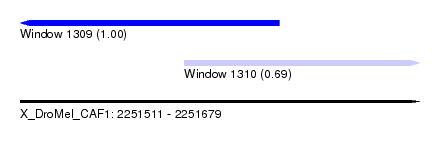
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,251,511 – 2,251,679 |
| Length | 168 |
| Max. P | 0.998603 |

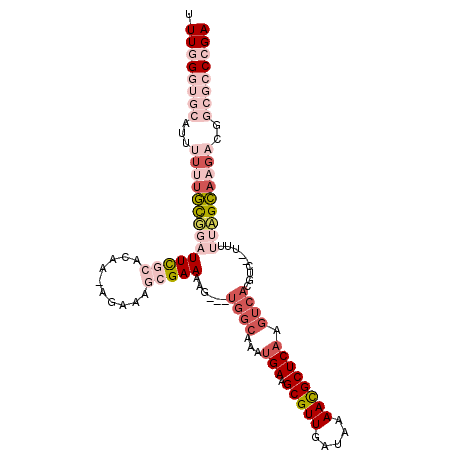
| Location | 2,251,511 – 2,251,620 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 74.17 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -15.42 |
| Energy contribution | -19.90 |
| Covariance contribution | 4.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2251511 109 - 22224390 UUUGGGUGCAUUUUUUGCGGAUUCGCACAAAAGAAAGCGAAAAGUAGUGGCAAAUGAAGCGUUGAUAAAACGCUCAAGUCACGUUUGUUUUUCGCAAGACGGCGCCCGA .((((((((...((((((((((((((..........))))).....(((((...(((.(((((.....)))))))).)))))........)))))))))..)))))))) ( -42.60) >DroPse_CAF1 16823 89 - 1 UUUGGGUGCAUUUUUUUUUUUUUUU---------UGGCGAAAAG---UGGCAAAUGAAGCGUUGAUAAAAUGCUCAAGUCACGUC---UUUUAGAAAGUC-----CAGA ((((((.((................---------..))((...(---((((...(((.(((((.....)))))))).))))).))---..........))-----)))) ( -16.67) >DroSec_CAF1 14396 106 - 1 UUUGGGUGCAUUUUUUGCGGAUUCGCACAAAAGAAAGCGAAAAG---UGGCAAAUGAAGCGUUGAUAAAACGCUCAAGUCACGUUUGUUUUUCGCAAGACGGCGCCCGA .((((((((...((((((((((((((..........)))))..(---((((...(((.(((((.....)))))))).)))))........)))))))))..)))))))) ( -42.60) >DroYak_CAF1 15364 93 - 1 UUUGGGUGCAUUUUCUGCGGAUUCGCACAA-AGAAAGCGAAAAG---UGGCAAAUGAAGCGUUGAUAAAACGCUC------------UUUUUGGCAAGACGGCGCCCGA .((((((((....(((.....(((((....-.....)))))...---((.((((...((((((.....)))))).------------..)))).)))))..)))))))) ( -31.60) >DroPer_CAF1 17417 84 - 1 UUUGGGUGCAU-----UUUUUUUUU---------UGGCGAAAAG---UGGCAAAUGAAGCGUUGAUAAAAUGCUCAAGUCACGUC---AUUUAGAAAGUC-----CAGA (((((....((-----(((((....---------((((.....(---((((...(((.(((((.....)))))))).))))))))---)...))))))))-----)))) ( -18.40) >consensus UUUGGGUGCAUUUUUUGCGGAUUCGCACAA_AGAAAGCGAAAAG___UGGCAAAUGAAGCGUUGAUAAAACGCUCAAGUCACGUC__UUUUUAGCAAGACGGCGCCCGA .((((((((...((((((((((((((..........)))))......((((...(((.(((((.....)))))))).)))).........)))))))))..)))))))) (-15.42 = -19.90 + 4.48)

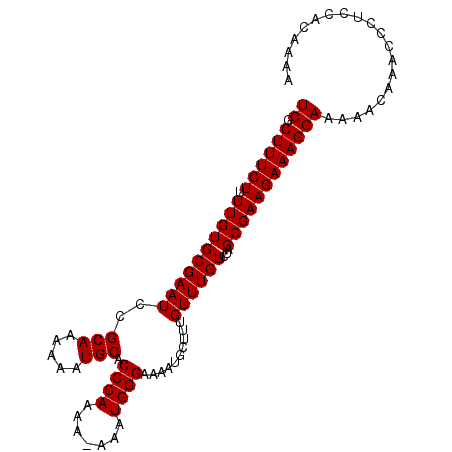
| Location | 2,251,580 – 2,251,679 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 94.65 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -18.57 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2251580 99 + 22224390 UCGCUUUCUUUUGUGCGAAUCCGCAAAAAAUGCACCCAAAA-AAAUGGGAAAAUGCUUUGUUUGUCUAGCGAAGAAAGGAAAAACAAACCCUCCACAAAC ((((..........)))).((((((.....))).((((...-...))))......(((((((.....)))))))...))).................... ( -19.90) >DroSec_CAF1 14462 98 + 1 UCGCUUUCUUUUGUGCGAAUCCGCAAAAAAUGCACCCAAA--AAAUGGGAAAAUGCUUUGUUUGUCUAGCGAAGAAAGGAAAAACAAACCCUCCACAAAA ((((..........)))).((((((.....))).((((..--...))))......(((((((.....)))))))...))).................... ( -19.70) >DroYak_CAF1 15418 99 + 1 UCGCUUUCU-UUGUGCGAAUCCGCAGAAAAUGCACCCAAAAAAAGUGGGAAAAUGCAUUGUUUGUCUAGCGAAGAAAGGAAAAACAAACCCUGCACAAAA ((.((((((-(((((((....)))(((.((((((((((.......))))....)))))).....))).)))))))))))).................... ( -26.40) >consensus UCGCUUUCUUUUGUGCGAAUCCGCAAAAAAUGCACCCAAAA_AAAUGGGAAAAUGCUUUGUUUGUCUAGCGAAGAAAGGAAAAACAAACCCUCCACAAAA ((.((((((.((((((((((..(((.....))).((((.......))))..........))))))...)))))))))))).................... (-18.57 = -18.57 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:19 2006