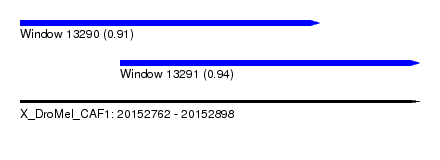
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,152,762 – 20,152,898 |
| Length | 136 |
| Max. P | 0.943419 |

| Location | 20,152,762 – 20,152,864 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.29 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -15.92 |
| Energy contribution | -16.03 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
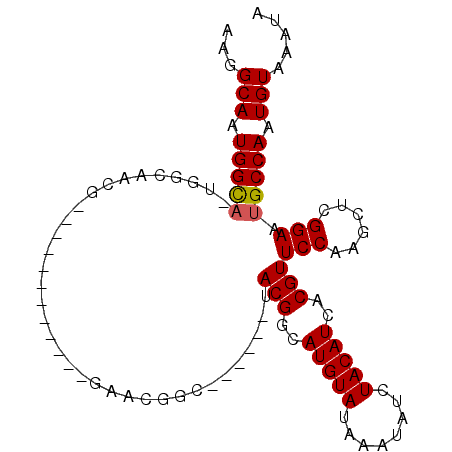
>X_DroMel_CAF1 20152762 102 + 22224390 AAGGCAAUGGCAAUGGCAACGGCAACGGCAGCGGAACGGCUACGACUACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCUCGGAAUGCCAAUGUAAAUA ...(((.(((((.((((..((....)).....(((((((((........)))(((((........)))))..))))))..)).))...))))).)))..... ( -30.10) >DroSec_CAF1 7806 84 + 1 AAGGCAAUGGCAGUGGCAACG------------GAACGGC------UACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCUCGGAAUGCCAAUGUAAAUA ...(((.(((((..(((...(------------(((((((------....))(((((........)))))..))))))..))).....))))).)))..... ( -27.10) >DroYak_CAF1 5951 78 + 1 AAGGCAAUGGU------AACG------------GAACGGC------UACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCUCGGAAUGCCAAUGUAAAUA ..((((.((((------...(------------(((((((------....))(((((........)))))..))))))..)).))...)))).......... ( -21.60) >consensus AAGGCAAUGGCA_UGGCAACG____________GAACGGC______UACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCUCGGAAUGCCAAUGUAAAUA ...(((.(((((...................................(((..(((((........)))))..)))(((......))).))))).)))..... (-15.92 = -16.03 + 0.11)


| Location | 20,152,796 – 20,152,898 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.95 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.94 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
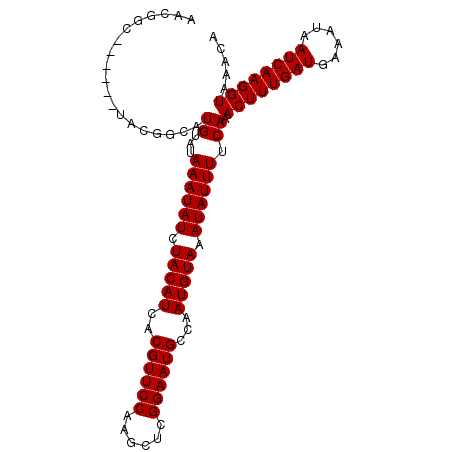
>X_DroMel_CAF1 20152796 102 + 22224390 AACGGCUACGACUACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCUCGGAAUGCCAAUGUAAAUAUUUUCAAACUUUGAUGAAAUAAUCAAGGUAAACA ....(((........))).((...((((((.(((((..((((((......))))))...))))).)))))).)).((((((((......))))))))..... ( -23.30) >DroSec_CAF1 7828 96 + 1 AACGGC------UACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCUCGGAAUGCCAAUGUAAAUAUUUUCAAACUUUGAUGAAAUAAUCAAGGUAAACA ....((------....)).((...((((((.(((((..((((((......))))))...))))).)))))).)).((((((((......))))))))..... ( -25.20) >DroEre_CAF1 3412 92 + 1 ----GC------UACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCUCGGAAUGCCAAUGUAAAUAUUUUCAAACUUUGAUGAAAUAAUCAAGGUAAACA ----((------....)).((...((((((.(((((..((((((......))))))...))))).)))))).)).((((((((......))))))))..... ( -25.20) >DroYak_CAF1 5967 96 + 1 AACGGC------UACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCUCGGAAUGCCAAUGUAAAUAUUUUCAAACUUUGAUGAAAUAAUCAAGGUAAACA ....((------....)).((...((((((.(((((..((((((......))))))...))))).)))))).)).((((((((......))))))))..... ( -25.20) >consensus AACGGC______UACGGCAUGUAUAAAUAUCUACAUCACGUUCCAAGCUCGGAAUGCCAAUGUAAAUAUUUUCAAACUUUGAUGAAAUAAUCAAGGUAAACA ...................((...((((((.(((((..((((((......))))))...))))).)))))).)).((((((((......))))))))..... (-21.40 = -21.40 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:22 2006