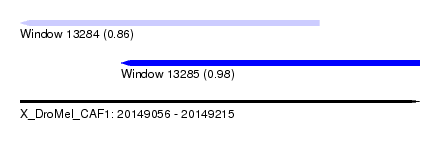
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,149,056 – 20,149,215 |
| Length | 159 |
| Max. P | 0.984415 |

| Location | 20,149,056 – 20,149,175 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.25 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -30.52 |
| Energy contribution | -31.32 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
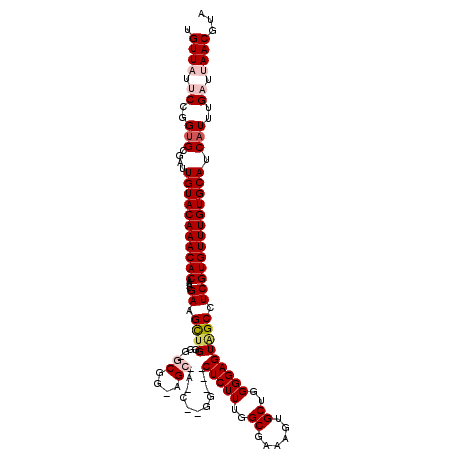
>X_DroMel_CAF1 20149056 119 - 22224390 GG-GCGGAAGCAACCGGGGGGACUCUUUGGCCAAAGUGCUGGGGAGUAGCCUCGUGUUUGUGCAUCAUUUGAUUAACGUAAUCGAAUGACACGCCACAUGUCAGUCAUCCAGAGAGGCAG .(-(((...((((((((((..((((((..((......))..))))))..))))).))...))).(((((((((((...)))))))))))..))))...((((..((.....))..)))). ( -45.80) >DroSec_CAF1 4334 101 - 1 GG-G------------------CUCUUUGGCGAAAGUGCUGGGGAGUAGCCUCGUGUUUGUGCAUCAUUUGAUUAACGUAAUCGAAUGACACGCCACAUGUCAGUCAUCCAGAGAGGCAG ..-(------------------(((((..(((....)))..)))))).((((((((..((((..(((((((((((...))))))))))))))).)))(((.....))).....))))).. ( -37.00) >DroSim_CAF1 9377 101 - 1 GG-G------------------CUCUUUGGCGAAAGUGCUGGGGAGUAGCCUCGUGUUUGUGCAUCAUUUGAUUAACGUAAUCGAAUGACACGCCACAUGUCAGUCAUCCAGAGAGGCAG ..-(------------------(((((..(((....)))..)))))).((((((((..((((..(((((((((((...))))))))))))))).)))(((.....))).....))))).. ( -37.00) >DroEre_CAF1 252 109 - 1 --GGCGGGAGCAACCAAGG---CUCUUUGGCCAAAGUGCUGGGGAGUAGCCUCGUGUUUGUGCAUCAUUUGAUUAACGUAAUCGAAUGACACGCCACAUGUCAGUCAUCCAGAG------ --(((((......))...(---(((((..((......))..)))))).))).(((((..(((..(((((((((((...)))))))))))..))).)))))..............------ ( -35.80) >DroYak_CAF1 2205 117 - 1 GGGGCGGGAGCGAGCAAGG---CUCUUUGGCGAAAGUGCUGGGGAGUGGCCUCGUGUUUGUGCAUCAUUUGAUUAACGUAAUCGAAUGACACGCCACAUGUCAGUCAUCCAGAGAGGCAG ...((....))......(.---(((((((((....))(((((.(.(((((....(((....)))(((((((((((...)))))))))))...))))).).)))))....))))))).).. ( -41.40) >consensus GG_GCGG_AGC_A_C__GG___CUCUUUGGCGAAAGUGCUGGGGAGUAGCCUCGUGUUUGUGCAUCAUUUGAUUAACGUAAUCGAAUGACACGCCACAUGUCAGUCAUCCAGAGAGGCAG ......................(((((..((......))..)))))..((((((((..((((..(((((((((((...))))))))))))))).)))(((.....))).....))))).. (-30.52 = -31.32 + 0.80)


| Location | 20,149,096 – 20,149,215 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.91 |
| Mean single sequence MFE | -40.84 |
| Consensus MFE | -30.22 |
| Energy contribution | -31.64 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20149096 119 - 22224390 UGUUAUUCCGGUGCGAUUGUACAAACACAAAUGAAGUUGGGG-GCGGAAGCAACCGGGGGGACUCUUUGGCCAAAGUGCUGGGGAGUAGCCUCGUGUUUGUGCAUCAUUUGAUUAACGUA .((((.((..(((....(((((((((((..........((..-((....))..))((((..((((((..((......))..))))))..))))))))))))))).)))..)).))))... ( -52.20) >DroSec_CAF1 4374 101 - 1 UGUUUUUCCGGUGCGAUUGUACAAACACAAAUGAAGCUGGGG-G------------------CUCUUUGGCGAAAGUGCUGGGGAGUAGCCUCGUGUUUGUGCAUCAUUUGAUUAACGUA .(((..((..(((....(((((((((((..........((((-(------------------(((((..(((....)))..))))))..))))))))))))))).)))..))..)))... ( -35.80) >DroSim_CAF1 9417 101 - 1 UGUUUUUCCGGUGCGAUUGUACAAACACAAAUGAAGCUGGGG-G------------------CUCUUUGGCGAAAGUGCUGGGGAGUAGCCUCGUGUUUGUGCAUCAUUUGAUUAACGUA .(((..((..(((....(((((((((((..........((((-(------------------(((((..(((....)))..))))))..))))))))))))))).)))..))..)))... ( -35.80) >DroEre_CAF1 286 115 - 1 UGUUAUCCUGGCGCGAUUGUACAAACACAAAUGAAGCCGG--GGCGGGAGCAACCAAGG---CUCUUUGGCCAAAGUGCUGGGGAGUAGCCUCGUGUUUGUGCAUCAUUUGAUUAACGUA .((((..(......(((.((((((((((....(....)((--(((((......))...(---(((((..((......))..)))))).))))))))))))))))))....)..))))... ( -39.90) >DroYak_CAF1 2245 117 - 1 UGUUAUCCCGGUGCGAUUGUACAAACACAAAUGAAGCUGGGGGGCGGGAGCGAGCAAGG---CUCUUUGGCGAAAGUGCUGGGGAGUGGCCUCGUGUUUGUGCAUCAUUUGAUUAACGUA .((((..(..(((....(((((((((((...............((....))((((...(---(((((..(((....)))..)))))).).)))))))))))))).)))..)..))))... ( -40.50) >consensus UGUUAUUCCGGUGCGAUUGUACAAACACAAAUGAAGCUGGGG_GCGG_AGC_A_C__GG___CUCUUUGGCGAAAGUGCUGGGGAGUAGCCUCGUGUUUGUGCAUCAUUUGAUUAACGUA .((((.((..(((....(((((((((((....((.((((....((....))...........(((((..((......))..))))))))).))))))))))))).)))..)).))))... (-30.22 = -31.64 + 1.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:17 2006