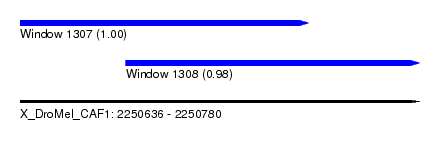
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,250,636 – 2,250,780 |
| Length | 144 |
| Max. P | 0.996685 |

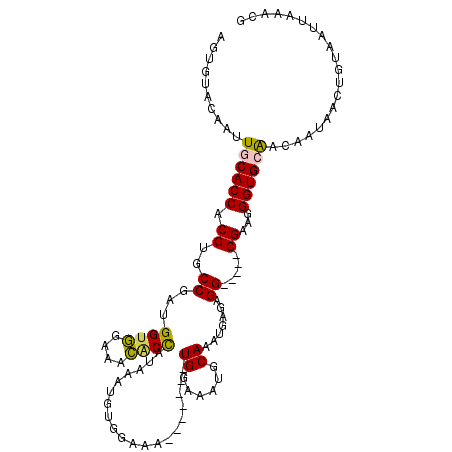
| Location | 2,250,636 – 2,250,740 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -24.49 |
| Energy contribution | -24.93 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2250636 104 + 22224390 U-CGAAAAAACGAUUAUUGUG-GUAAUUGGCAUUUUGUUCAGUGUACAAUUGCACCACGUGCCGAUGGUGGAAACACUAUAAAUGUGGAAA-------UGGAAAUGCAAAUGC .-........(((((((....-)))))))(((((((.(((.(((((....)))))(((((....((((((....))))))..)))))))).-------..)))))))...... ( -31.20) >DroSec_CAF1 13500 106 + 1 UUUGGUAAAACAAUCAUUGCGGGUAAGCGGCAUUUUGUUCAGUGUACAAUUGCACCACGUGCCGAUGGAGGAAACACCAUAAAUGUGGAAA-------UGGAAAUGCAAAUGC .(((......))).((((((......)).(((((((.(((.(((((....)))))(((((....((((.(....).))))..)))))))).-------..))))))).)))). ( -27.30) >DroYak_CAF1 14528 101 + 1 ------------CUCAAUGUGGCAACUUGGCCUUUUGUUCAGUGUACAAUUGCACCACGUGCCGAUGGUGGAAACACCAUAAAUGUGGAAAUGUAUAAUGGAAAUGCAAAUGU ------------.....((((((......))).....((((.((((((.((...((((((....((((((....))))))..)))))).)))))))).))))...)))..... ( -31.30) >consensus U__G__AAAAC_AUCAUUGUGGGUAAUUGGCAUUUUGUUCAGUGUACAAUUGCACCACGUGCCGAUGGUGGAAACACCAUAAAUGUGGAAA_______UGGAAAUGCAAAUGC .............................(((((((...((((.....))))..((((((....((((((....))))))..))))))............)))))))...... (-24.49 = -24.93 + 0.45)

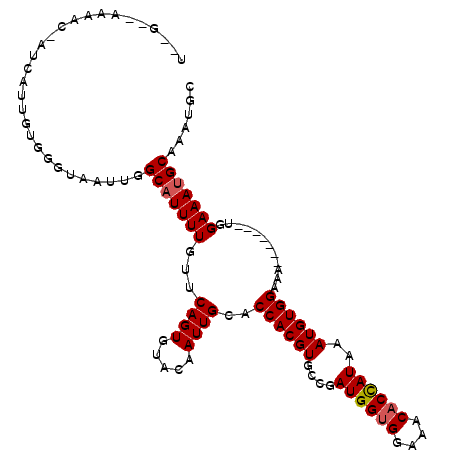
| Location | 2,250,674 – 2,250,780 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.21 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -13.46 |
| Energy contribution | -13.02 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2250674 106 + 22224390 AGUGUACAAUUGCACCACGUGCCGAUGGUGGAAACACUAUAAAUGUGGAAA-------UGGAAAUGCAAAUGCGAGGCGGGGCGAAGGGUGCAACAAUAACUGUAAUUAAACG (((......(((((((.(((.(((((((((....))))))...........-------......(((....)))...))).)))...))))))).....)))........... ( -32.30) >DroPse_CAF1 15859 85 + 1 ---------GUACACCACGUGCCCAAGGUUGGCUGGCCG-------GUAAA-------UGCAAAUGCAAAUGAGAGG-----CGAAGGGUGCGACAAUAACUGUAAUUAAACG ---------........((..(((..((....)).(((.-------.((..-------(((....)))..))...))-----)...)))..)).................... ( -19.50) >DroSec_CAF1 13540 106 + 1 AGUGUACAAUUGCACCACGUGCCGAUGGAGGAAACACCAUAAAUGUGGAAA-------UGGAAAUGCAAAUGCGAGGCGGGGCGAAGGGUGCAACAAUAACUGUAAUUAAAAG (((......(((((((.(((.(((((((.(....).))))...........-------......(((....)))...))).)))...))))))).....)))........... ( -29.90) >DroYak_CAF1 14556 108 + 1 AGUGUACAAUUGCACCACGUGCCGAUGGUGGAAACACCAUAAAUGUGGAAAUGUAUAAUGGAAAUGCAAAUGUGAGG-----CGAAGGGUGCAACAAUAACUGUAAUUAAACG (((......(((((((...((((.((((((....))))))...........(((((.......))))).......))-----))...))))))).....)))........... ( -33.10) >DroPer_CAF1 16395 85 + 1 ---------GUACACCACGUGCCCAAGGUUGGCUGGCCG-------GUAAA-------UGCAAAUGCAAAUGAGAGG-----CGAAGGGUGCGACAAUAACAGUAAUUAAACG ---------........((..(((..((....)).(((.-------.((..-------(((....)))..))...))-----)...)))..)).................... ( -19.50) >consensus AGUGUACAAUUGCACCACGUGCCGAUGGUGGAAACACCAUAAAUGUGGAAA_______UGGAAAUGCAAAUGAGAGG_____CGAAGGGUGCAACAAUAACUGUAAUUAAACG ..........((((((.((..((...((((....))))....................((......)).......)).....))...)))))).................... (-13.46 = -13.02 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:17 2006