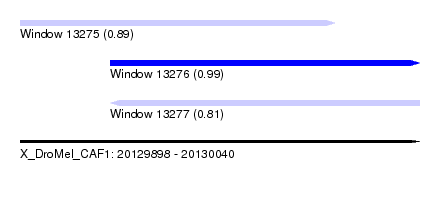
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,129,898 – 20,130,040 |
| Length | 142 |
| Max. P | 0.988905 |

| Location | 20,129,898 – 20,130,010 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -22.72 |
| Energy contribution | -23.17 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20129898 112 + 22224390 ------UUAGUCCACAGAAUGCCACAGAAGUGCAUG--UGGUAUUGAAAUCACAAGAACCUAAAAGUAUGCAGUGAAAAUACAAUUUAAAAUGUAACGCAGCAUACUUUUACACACCUUU ------...........(((((((((........))--)))))))...............(((((((((((.(((....((((........)))).))).)))))))))))......... ( -26.70) >DroSec_CAF1 31367 120 + 1 UUUCCACUAGUCCACAGAAUGCCACAGAAGUGCAUGUGUGGUGUUGAAAUCACAAGAACCUGAAAGUAUGCAAUGAAAAUACAUUCUAAAAUGUAGCGCAGCAUACUUUUACACACCCUU ((((.((....(((((..((((.((....)))))).))))).)).))))...........(((((((((((..((....((((((....)))))).))..)))))))))))......... ( -30.30) >DroSim_CAF1 14828 114 + 1 ------CUAGUCCACAGAAUGCCACAGAAGUGCAUGUGUGGUGUUGAAAUCACAAGAACCUGAAAGUAUGCAAUGAAAAUACAUUCUAAAAUGUAACGCAGCAUACUUUUACACACCCUU ------............((((.((....))))))((((((((.......))).......(((((((((((..((....((((((....))))))...)))))))))))))))))).... ( -26.80) >consensus ______CUAGUCCACAGAAUGCCACAGAAGUGCAUGUGUGGUGUUGAAAUCACAAGAACCUGAAAGUAUGCAAUGAAAAUACAUUCUAAAAUGUAACGCAGCAUACUUUUACACACCCUU ...........((((...((((.((....))))))..))))...................(((((((((((..((....((((((....))))))...)))))))))))))......... (-22.72 = -23.17 + 0.45)

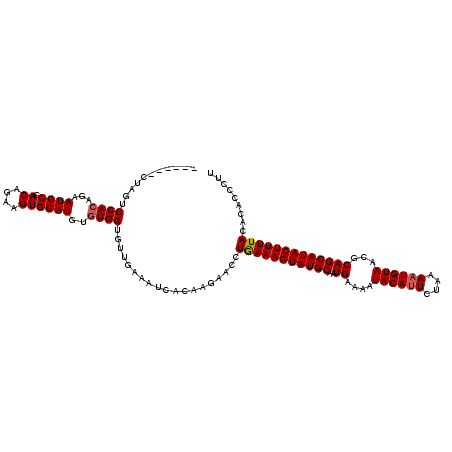
| Location | 20,129,930 – 20,130,040 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.12 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -24.23 |
| Energy contribution | -24.34 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20129930 110 + 22224390 GUAUUGAAAUCACAAGAACCUAAAAGUAUGCAGUGAAAAUACAAUUUAAAAUGUAACGCAGCAUACUUUUACACACCUUUAUACAUACAUGUGUGCUAUAUUGUGUUCAU ....((((..(((((.....(((((((((((.(((....((((........)))).))).)))))))))))(((((..............))))).....))))))))). ( -24.54) >DroSec_CAF1 31407 106 + 1 GUGUUGAAAUCACAAGAACCUGAAAGUAUGCAAUGAAAAUACAUUCUAAAAUGUAGCGCAGCAUACUUUUACACACCCUUAUA----CAUGUGUGAUAUGUUGUGUUCAU ....((((..(((((.(...(((((((((((..((....((((((....)))))).))..)))))))))))(((((.......----...)))))...).))))))))). ( -27.00) >DroSim_CAF1 14862 110 + 1 GUGUUGAAAUCACAAGAACCUGAAAGUAUGCAAUGAAAAUACAUUCUAAAAUGUAACGCAGCAUACUUUUACACACCCUUAUACAUACAUGUGUGAUAUGUUGUGUUCAU ....((((..(((((.(...(((((((((((..((....((((((....))))))...)))))))))))))(((((..............)))))...).))))))))). ( -25.34) >consensus GUGUUGAAAUCACAAGAACCUGAAAGUAUGCAAUGAAAAUACAUUCUAAAAUGUAACGCAGCAUACUUUUACACACCCUUAUACAUACAUGUGUGAUAUGUUGUGUUCAU ....((((..(((((.....(((((((((((..((....((((((....))))))...)))))))))))))(((((..............))))).....))))))))). (-24.23 = -24.34 + 0.11)


| Location | 20,129,930 – 20,130,040 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.12 |
| Mean single sequence MFE | -25.81 |
| Consensus MFE | -22.13 |
| Energy contribution | -23.02 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20129930 110 - 22224390 AUGAACACAAUAUAGCACACAUGUAUGUAUAAAGGUGUGUAAAAGUAUGCUGCGUUACAUUUUAAAUUGUAUUUUCACUGCAUACUUUUAGGUUCUUGUGAUUUCAAUAC .(((((((((....((((((...((....))...))))))((((((((((.(.(.((((........))))....).).))))))))))......)))))..)))).... ( -26.50) >DroSec_CAF1 31407 106 - 1 AUGAACACAACAUAUCACACAUG----UAUAAGGGUGUGUAAAAGUAUGCUGCGCUACAUUUUAGAAUGUAUUUUCAUUGCAUACUUUCAGGUUCUUGUGAUUUCAACAC .(((((((((...(((((((((.----(....).)))))).(((((((((.....((((((....))))))........)))))))))..)))..)))))..)))).... ( -25.12) >DroSim_CAF1 14862 110 - 1 AUGAACACAACAUAUCACACAUGUAUGUAUAAGGGUGUGUAAAAGUAUGCUGCGUUACAUUUUAGAAUGUAUUUUCAUUGCAUACUUUCAGGUUCUUGUGAUUUCAACAC .(((((((((...(((((((((.(........).)))))).(((((((((.....((((((....))))))........)))))))))..)))..)))))..)))).... ( -25.82) >consensus AUGAACACAACAUAUCACACAUGUAUGUAUAAGGGUGUGUAAAAGUAUGCUGCGUUACAUUUUAGAAUGUAUUUUCAUUGCAUACUUUCAGGUUCUUGUGAUUUCAACAC .(((((((((...(((((((((.(........).)))))).(((((((((.....((((((....))))))........)))))))))..)))..)))))..)))).... (-22.13 = -23.02 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:09 2006