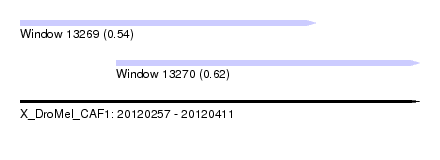
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,120,257 – 20,120,411 |
| Length | 154 |
| Max. P | 0.617492 |

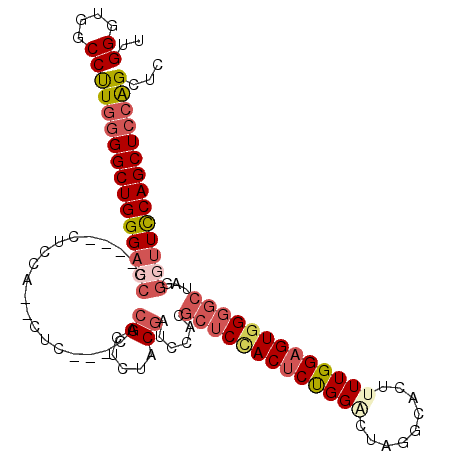
| Location | 20,120,257 – 20,120,371 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -45.46 |
| Consensus MFE | -28.74 |
| Energy contribution | -31.30 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20120257 114 + 22224390 UUGGGUGGCCUUGGGGCUGGGACCGCUUCCUCCACUCUCGCUCACGCUCUACGAUCCACGCUCCACUCUGGCCUAGGCACUUUUGGAGUGGGGCUAGGGUUCCAGCUCCAGCUC ......(((..(((((((((((((.((..(((((((((.((....))............(((((.....))....)))......)))))))))..)))))))))))))))))). ( -47.40) >DroSec_CAF1 22123 107 + 1 UUGGGUGGCCUUGGGGCUGGG----CUCCCUCCACGCUC---CACGCUCUACGAUCCAUGCUCUACUCUGGGCUAGGCACUUUUGGUGUGGGGCUAGGGUUCCAGCUCCAGCUC ......(((..((((((((((----..((((....((((---((((((.......((..((((......))))..)).......)))))))))).)))).))))))))))))). ( -48.54) >DroEre_CAF1 17025 98 + 1 UUGGGUGGCCCUGGGGCUGGGAGCG----CUCCC--C-----CACGCUCUCCAAUCCCCGCUCCACUCUGGACUAGGCGCUGUUGGAGUUG-----GGGUUCCAGCUGGGGUUC ......((((((..(((((((((((----.....--.-----..)))))...((((((.(((((((..((.......))..).)))))).)-----))))))))))))))))). ( -40.50) >DroYak_CAF1 1052 105 + 1 UUGGGUGGCCUUGGGGCUGGGACCG----CUCCC--CUU---CACGCUCUCCGAUUUACGCUCCACUCCGGACUAGGUACUUUUGGAGUGGGGCUAGGGUUUCAGCUCCAGCUC ......(((..((((((((..((((----(....--...---...))............(((((((((((((.........)))))))))))))...)))..))))))))))). ( -45.40) >consensus UUGGGUGGCCUUGGGGCUGGGACCG____CUCCA__CUC___CACGCUCUACGAUCCACGCUCCACUCUGGACUAGGCACUUUUGGAGUGGGGCUAGGGUUCCAGCUCCAGCUC ..((....))((((((((((((((....................((.....))......(((((((((((((.........)))))))))))))...))))))))))))))... (-28.74 = -31.30 + 2.56)


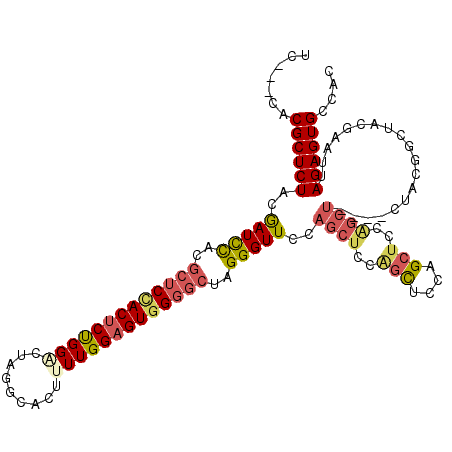
| Location | 20,120,294 – 20,120,411 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.65 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -23.27 |
| Energy contribution | -25.15 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20120294 117 + 22224390 UCGCUCACGCUCUACGAUCCACGCUCCACUCUGGCCUAGGCACUUUUGGAGUGGGGCUAGGGUUCCAGCUCCAGCUCCAGCUCCAGCUAUAGCUACGGCUACGAAUUAGAGUGCCAC .......(((((((.(((((..((((((((((.((....))......))))))))))..)))))..((((..(((....)))..)))).((((....)))).....))))))).... ( -43.70) >DroSec_CAF1 22156 102 + 1 UC---CACGCUCUACGAUCCAUGCUCUACUCUGGGCUAGGCACUUUUGGUGUGGGGCUAGGGUUCCAGCUCCAGCUCCGG------------CUACGACUACGAAUUAGAGUGCCAC ..---.................((((......))))..((((((((((((.(((((((.((((....)))).))))))))------------)))((....))....)))))))).. ( -32.70) >DroEre_CAF1 17056 101 + 1 -----CACGCUCUCCAAUCCCCGCUCCACUCUGGACUAGGCGCUGUUGGAGUUG-----GGGUUCCAGCUGGGGUUCCAGCUCCAGCU------ACCGCUACGAAUUAGAGUGCCAC -----..((((((...........(((.....)))...((((..((((((((((-----(((..((....))..))))))))))))).------..)))).......)))))).... ( -41.50) >DroYak_CAF1 1083 114 + 1 UU---CACGCUCUCCGAUUUACGCUCCACUCCGGACUAGGUACUUUUGGAGUGGGGCUAGGGUUUCAGCUCCAGCUCCAGCUACGGCUACGACUACGGCUACGAAUUAGAGUGCCAC ..---...((.(((..((((..(((((((((((((.........)))))))))))))...((((..((((..(((....)))..))))..))))........))))..))).))... ( -40.10) >consensus UC___CACGCUCUACGAUCCACGCUCCACUCUGGACUAGGCACUUUUGGAGUGGGGCUAGGGUUCCAGCUCCAGCUCCAGCUCCAGCU____CUACGGCUACGAAUUAGAGUGCCAC .......((((((..(((((..(((((((((((((.........)))))))))))))..)))))..((((..(((....)))..))))...................)))))).... (-23.27 = -25.15 + 1.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:03 2006