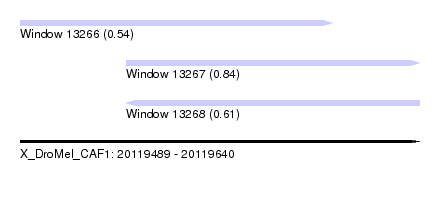
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,119,489 – 20,119,640 |
| Length | 151 |
| Max. P | 0.837180 |

| Location | 20,119,489 – 20,119,607 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -45.44 |
| Consensus MFE | -36.49 |
| Energy contribution | -37.43 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20119489 118 + 22224390 AGCGCCAAAGAGCAAAGUGCGCCUACAACAUGGCCAUCCCACCAAGUAGGAAUCCAAUGGGGCGGUGGUCUGCAGCCCACGGGCCGUGUGCCAUGGGCCCAACUGGGCGUGGCAGCAG .((((((....((..(((..(((((..((((((((.(((.((...)).)))......((((((((....))))..))))..))))))))....)))))...)))..)).)))).)).. ( -48.10) >DroSec_CAF1 21336 118 + 1 AGCGCCAAAGAGCAAAGUGCGCGUACAACAUGUCCAACCCACCAAGUAGGCAUCCAGUGGGGCGGUGGCCUGCAGCCCACGGGCCGUGUGCCAUGGGCCCAACUGGGCGUGGCAGCAG .((((((....((.......((.(((...................))).))..((((((((.(.(((((.(((.(((....))).))).))))).).))).))))))).)))).)).. ( -49.61) >DroYak_CAF1 260 94 + 1 AGCGCCAAAGAGCAAAGUGCGCCUACAACAUGC----UCCACCAAGCAGGAAUCCAGUGGGGGGGCGG--------------------UGCCGUGGGCCCAACUGGGCGUGGCAGCAG .((((((....((..(((..((((((.((.(((----(((.(((....((...))..))).)))))))--------------------)...))))))...)))..)).)))).)).. ( -38.60) >consensus AGCGCCAAAGAGCAAAGUGCGCCUACAACAUGCCCA_CCCACCAAGUAGGAAUCCAGUGGGGCGGUGG_CUGCAGCCCACGGGCCGUGUGCCAUGGGCCCAACUGGGCGUGGCAGCAG .((((((....((.....))((...................((.....))...((((((((.(.(((((.(((.(((....))).))).))))).).))).))))))).)))).)).. (-36.49 = -37.43 + 0.95)


| Location | 20,119,529 – 20,119,640 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -38.29 |
| Energy contribution | -39.23 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20119529 111 + 22224390 ACCAAGUAGGAAUCCAAUGGGGCGGUGGUCUGCAGCCCACGGGCCGUGUGCCAUGGGCCCAACUGGGCGUGGCAGCAGGACAAAAA--GGGAACGUGUUAAGCCGGCCACAUU .((.....))..((((.((((.(.(((((..((.(((....))).))..))))).).))))..)))).(((((.((..((((....--(....).))))..))..)))))... ( -45.50) >DroSec_CAF1 21376 111 + 1 ACCAAGUAGGCAUCCAGUGGGGCGGUGGCCUGCAGCCCACGGGCCGUGUGCCAUGGGCCCAACUGGGCGUGGCAGCAGGACAAAAA--GGGAACGUGUUAAGCCGGCCACAUU .........((..((((((((.(.(((((.(((.(((....))).))).))))).).))).)))))))(((((.((..((((....--(....).))))..))..)))))... ( -50.80) >DroYak_CAF1 296 93 + 1 ACCAAGCAGGAAUCCAGUGGGGGGGCGG--------------------UGCCGUGGGCCCAACUGGGCGUGGCAGCAGGACAAGAACGGGGAACGUGUUAAGCCGGCCACAUU .((.....))..((((((.(((..(((.--------------------...)))...))).)))))).(((((....((......(((.....)))......)).)))))... ( -32.40) >consensus ACCAAGUAGGAAUCCAGUGGGGCGGUGG_CUGCAGCCCACGGGCCGUGUGCCAUGGGCCCAACUGGGCGUGGCAGCAGGACAAAAA__GGGAACGUGUUAAGCCGGCCACAUU .((.....))..(((((((((.(.(((((.(((.(((....))).))).))))).).))).)))))).(((((.((..((((......(....).))))..))..)))))... (-38.29 = -39.23 + 0.95)


| Location | 20,119,529 – 20,119,640 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -33.07 |
| Energy contribution | -32.93 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20119529 111 - 22224390 AAUGUGGCCGGCUUAACACGUUCCC--UUUUUGUCCUGCUGCCACGCCCAGUUGGGCCCAUGGCACACGGCCCGUGGGCUGCAGACCACCGCCCCAUUGGAUUCCUACUUGGU ...(((((.(((...(((.......--....)))...))))))))..(((((.((((...(((....(((((....)))))....)))..)))).)))))...((.....)). ( -37.40) >DroSec_CAF1 21376 111 - 1 AAUGUGGCCGGCUUAACACGUUCCC--UUUUUGUCCUGCUGCCACGCCCAGUUGGGCCCAUGGCACACGGCCCGUGGGCUGCAGGCCACCGCCCCACUGGAUGCCUACUUGGU ...(((((.(((...(((.......--....)))...))))))))..(((((.((((...((((...(((((....)))))...))))..)))).)))))..(((.....))) ( -46.00) >DroYak_CAF1 296 93 - 1 AAUGUGGCCGGCUUAACACGUUCCCCGUUCUUGUCCUGCUGCCACGCCCAGUUGGGCCCACGGCA--------------------CCGCCCCCCCACUGGAUUCCUGCUUGGU ...(((((.(((...(((.(.........).)))...))))))))..(((((.(((....((...--------------------.))...))).)))))...((.....)). ( -26.10) >consensus AAUGUGGCCGGCUUAACACGUUCCC__UUUUUGUCCUGCUGCCACGCCCAGUUGGGCCCAUGGCACACGGCCCGUGGGCUGCAG_CCACCGCCCCACUGGAUUCCUACUUGGU ...(((((.(((...(((.(.........).)))...))))))))..(((((.((((...(((....(((((....)))))....)))..)))).)))))...((.....)). (-33.07 = -32.93 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:02 2006