
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,115,980 – 20,116,214 |
| Length | 234 |
| Max. P | 0.897024 |

| Location | 20,115,980 – 20,116,100 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -14.35 |
| Consensus MFE | -13.45 |
| Energy contribution | -13.45 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.94 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
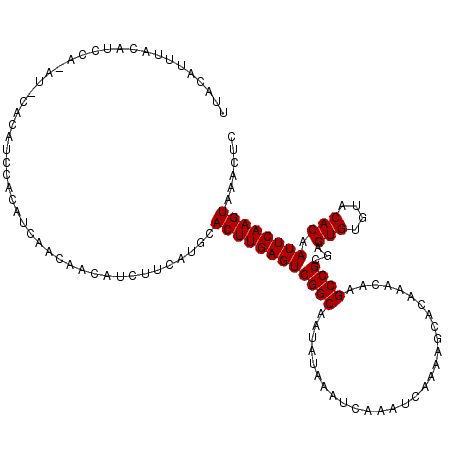
>X_DroMel_CAF1 20115980 120 - 22224390 UUACAUUUACAUUCACAUUCACAUCCACAUCAACAACAUCUUCAUGCACUUGAGUCGGCAAUAUAAAUCAAAUCAAAAGCACAAACAAGCCGCGAGUGUGUACACAAUUCAAGUAAACUC .............(((((((.........((((...(((....)))...))))(.((((.............................)))))))))))).................... ( -14.35) >DroSec_CAF1 16534 114 - 1 UUACAUUUACAUC------CGCAUCCACAUCAACAACAUCUUCAUGCACUUGAGUCGGCAAUAUAAAUCAAAUCAAAAGCACAAACAAGCCGCGAGUGUGUACACAAUUCAAGUAAACUC .............------.((((...................))))((((((((((((.............................))))...(((....))).))))))))...... ( -14.96) >DroEre_CAF1 12982 109 - 1 -----------UCCAUAUACACAAUCACAUCAACAACAUCUUCAUGCACUUGAGUCGGCAAUAUAAAUCAAAUCAAAAGCACAAACAAGCCGCGAGUGUGUACACAAUUCAAGUAAACUC -----------......((((((.((...((((...(((....)))...))))(.((((.............................))))))).)))))).................. ( -13.75) >consensus UUACAUUUACAUCCA_AU_CACAUCCACAUCAACAACAUCUUCAUGCACUUGAGUCGGCAAUAUAAAUCAAAUCAAAAGCACAAACAAGCCGCGAGUGUGUACACAAUUCAAGUAAACUC ...............................................((((((((((((.............................))))...(((....))).))))))))...... (-13.45 = -13.45 + 0.00)

| Location | 20,116,100 – 20,116,214 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.35 |
| Mean single sequence MFE | -18.70 |
| Consensus MFE | -14.09 |
| Energy contribution | -13.77 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20116100 114 - 22224390 CCAACAGCAGACAGAGAUCAUCAUCAUCA------UGGUGGCCCGAAAAUCGGAGCACAAGUUCUGGCCCACUCACAUCCACAUCCACAUCCAUAUCCACACUCGCACUCACACUCACAU ...........(((((..((((((....)------)))))(((((.....))).)).....)))))...................................................... ( -15.80) >DroSec_CAF1 16648 101 - 1 CCAACAGCAGACAGAGAUCAUCAUCAUCAUCAUCCUGGUGGCCCCA-AAUCGGAGCACAAGUUCUGGCCCACACACAUCCACAUCCA------UAUGCA------------CACUCACAU ......(((..(((.(((.((.......)).))))))((((.((..-....(((((....))))))).))))...............------..))).------------......... ( -16.80) >DroEre_CAF1 13091 100 - 1 CCGACACCAGACAGAGAUCAUCAUCAUCGUCGUCAUGGUGGACCCG-AACCGGAGCACAAGCUCUGGCCCACUCACAUCCACAUCCACAUGCACAUCC------------------ACA- .........(((.(((((....))).)))))(((((((((((...(-(.(((((((....))))))).....))...))))).....)))).))....------------------...- ( -23.50) >consensus CCAACAGCAGACAGAGAUCAUCAUCAUCAUC_UC_UGGUGGCCCCA_AAUCGGAGCACAAGUUCUGGCCCACUCACAUCCACAUCCACAU_CAUAUCCA____________CACUCACAU .............(((.......((((((......))))))........(((((((....)))))))....))).............................................. (-14.09 = -13.77 + -0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:58 2006