
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,103,700 – 20,103,851 |
| Length | 151 |
| Max. P | 0.974575 |

| Location | 20,103,700 – 20,103,815 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -44.87 |
| Consensus MFE | -39.25 |
| Energy contribution | -38.37 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20103700 115 + 22224390 GUUGUCCACCUGUAACCUGUUGAUAUGUACAGCCUGCAGUGCGGGCAAAGUGGGCGGAACGGAUGACCAUGGCUGCAUGCACCAGUAUAGGCCACCUGUAACGCCCACUUCCCAG .((((((.((((((..((((........))))..))))).).))))))((((((((..((((....)).(((((..((((....)))).)))))...))..))))))))...... ( -46.60) >DroSec_CAF1 13758 113 + 1 GUUGUCCACCUGUAACCUGUUGAUAUGUACAGCCUGCAGUGCGGGAAAAGUGGGCGGGACGGAUGACCAUGGCU--AUGCACCUGUCUAGGCCACCUGUAACGCCCACUUCCCAG ......(((.((((..((((........))))..))))))).((((..((((((((..((((.((.((..(((.--........)))..)).)).))))..)))))))))))).. ( -43.50) >DroEre_CAF1 10723 108 + 1 GUUAUCCACCUGUAACCUGUUGAUAUGUACAGCCUGCAGUGCGGGAAAAGUGGGCGGGACGGAUGACUAUAGCU--AUGUAUAU-----GGCCGCCCGUAACGCCCACUUCCCAG ......(((.((((..((((........))))..))))))).((((..((((((((..((((.((.(((((...--.....)))-----)).)).))))..)))))))))))).. ( -44.50) >consensus GUUGUCCACCUGUAACCUGUUGAUAUGUACAGCCUGCAGUGCGGGAAAAGUGGGCGGGACGGAUGACCAUGGCU__AUGCACCUGU_UAGGCCACCUGUAACGCCCACUUCCCAG ......(((.((((..((((........))))..))))))).(((..(((((((((..((((.((.((...((.....)).........)).)).))))..)))))))))))).. (-39.25 = -38.37 + -0.88)



| Location | 20,103,700 – 20,103,815 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -48.27 |
| Consensus MFE | -41.73 |
| Energy contribution | -42.40 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20103700 115 - 22224390 CUGGGAAGUGGGCGUUACAGGUGGCCUAUACUGGUGCAUGCAGCCAUGGUCAUCCGUUCCGCCCACUUUGCCCGCACUGCAGGCUGUACAUAUCAACAGGUUACAGGUGGACAAC ..(.((((((((((..((.(((((((.....((((.......)))).))))))).))..)))))))))).)((((.(((.((.((((........)))).)).)))))))..... ( -49.80) >DroSec_CAF1 13758 113 - 1 CUGGGAAGUGGGCGUUACAGGUGGCCUAGACAGGUGCAU--AGCCAUGGUCAUCCGUCCCGCCCACUUUUCCCGCACUGCAGGCUGUACAUAUCAACAGGUUACAGGUGGACAAC ..(..(((((((((..((.(((((((......(((....--.)))..))))))).))..)))))))))..)((((.(((.((.((((........)))).)).)))))))..... ( -49.00) >DroEre_CAF1 10723 108 - 1 CUGGGAAGUGGGCGUUACGGGCGGCC-----AUAUACAU--AGCUAUAGUCAUCCGUCCCGCCCACUUUUCCCGCACUGCAGGCUGUACAUAUCAACAGGUUACAGGUGGAUAAC ..(..(((((((((..((((..(((.-----(((.....--...))).)))..))))..)))))))))..)((((.(((.((.((((........)))).)).)))))))..... ( -46.00) >consensus CUGGGAAGUGGGCGUUACAGGUGGCCUA_ACAGGUGCAU__AGCCAUGGUCAUCCGUCCCGCCCACUUUUCCCGCACUGCAGGCUGUACAUAUCAACAGGUUACAGGUGGACAAC ..(..(((((((((..((.(((((((.....................))))))).))..)))))))))..)((((.(((.((.((((........)))).)).)))))))..... (-41.73 = -42.40 + 0.67)


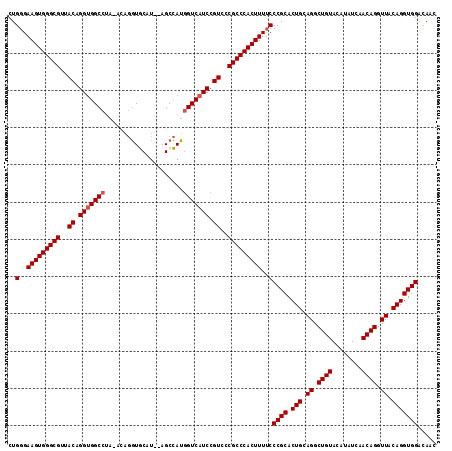
| Location | 20,103,740 – 20,103,851 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -37.05 |
| Energy contribution | -36.17 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20103740 111 + 22224390 GCGGGCAAAGUGGGCGGAACGGAUGACCAUGGCUGCAUGCACCAGUAUAGGCCACCUGUAACGCCCACUUCCCAGAUCUGUGAGUUUGUGGCAUUGCAAACGUUUUAUCAC ..(((..(((((((((..((((....)).(((((..((((....)))).)))))...))..)))))))))))).(((......((((((......)))))).....))).. ( -41.20) >DroSec_CAF1 13798 109 + 1 GCGGGAAAAGUGGGCGGGACGGAUGACCAUGGCU--AUGCACCUGUCUAGGCCACCUGUAACGCCCACUUCCCAGAUCUGUGAGUUUGUGGCAUUGCAAACGUUUUAUCAC ..((((..((((((((..((((.((.((..(((.--........)))..)).)).))))..)))))))))))).(((......((((((......)))))).....))).. ( -41.30) >DroEre_CAF1 10763 104 + 1 GCGGGAAAAGUGGGCGGGACGGAUGACUAUAGCU--AUGUAUAU-----GGCCGCCCGUAACGCCCACUUCCCAGAUCUGUGAGUUUGUGGCAUUGCAAACGUUUUAUCAC ..((((..((((((((..((((.((.(((((...--.....)))-----)).)).))))..)))))))))))).(((......((((((......)))))).....))).. ( -42.30) >consensus GCGGGAAAAGUGGGCGGGACGGAUGACCAUGGCU__AUGCACCUGU_UAGGCCACCUGUAACGCCCACUUCCCAGAUCUGUGAGUUUGUGGCAUUGCAAACGUUUUAUCAC ..(((..(((((((((..((((.((.((...((.....)).........)).)).))))..)))))))))))).(((......((((((......)))))).....))).. (-37.05 = -36.17 + -0.88)

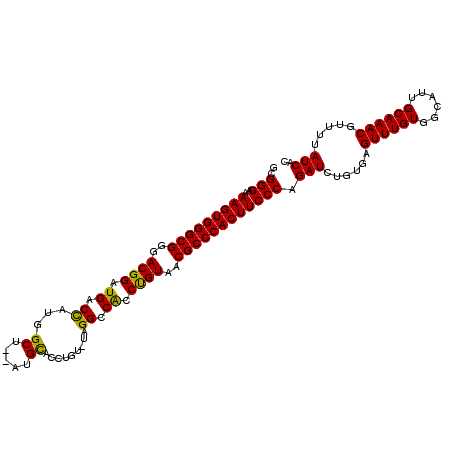
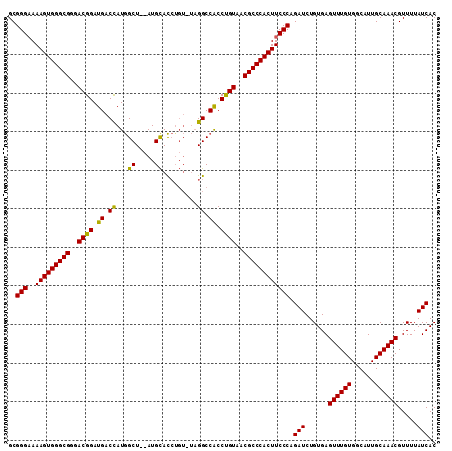
| Location | 20,103,740 – 20,103,851 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -41.07 |
| Consensus MFE | -34.60 |
| Energy contribution | -35.27 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20103740 111 - 22224390 GUGAUAAAACGUUUGCAAUGCCACAAACUCACAGAUCUGGGAAGUGGGCGUUACAGGUGGCCUAUACUGGUGCAUGCAGCCAUGGUCAUCCGUUCCGCCCACUUUGCCCGC ((((......(((((........)))))))))......(((.((((((((..((.(((((((.....((((.......)))).))))))).))..))))))))...))).. ( -42.80) >DroSec_CAF1 13798 109 - 1 GUGAUAAAACGUUUGCAAUGCCACAAACUCACAGAUCUGGGAAGUGGGCGUUACAGGUGGCCUAGACAGGUGCAU--AGCCAUGGUCAUCCGUCCCGCCCACUUUUCCCGC ((((......(((((........)))))))))......((((((((((((..((.(((((((......(((....--.)))..))))))).))..))))))))..)))).. ( -41.70) >DroEre_CAF1 10763 104 - 1 GUGAUAAAACGUUUGCAAUGCCACAAACUCACAGAUCUGGGAAGUGGGCGUUACGGGCGGCC-----AUAUACAU--AGCUAUAGUCAUCCGUCCCGCCCACUUUUCCCGC ((((......(((((........)))))))))......((((((((((((..((((..(((.-----(((.....--...))).)))..))))..))))))))..)))).. ( -38.70) >consensus GUGAUAAAACGUUUGCAAUGCCACAAACUCACAGAUCUGGGAAGUGGGCGUUACAGGUGGCCUA_ACAGGUGCAU__AGCCAUGGUCAUCCGUCCCGCCCACUUUUCCCGC ((((......(((((........)))))))))......((((((((((((..((.(((((((.....................))))))).))..)))))))))..))).. (-34.60 = -35.27 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:52 2006