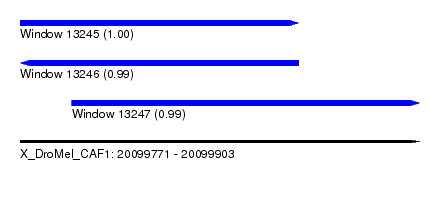
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,099,771 – 20,099,903 |
| Length | 132 |
| Max. P | 0.998054 |

| Location | 20,099,771 – 20,099,863 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -19.05 |
| Energy contribution | -18.67 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20099771 92 + 22224390 ----------------------UGUUUUG-UUUCGUGUUCAUCGCUUAAGCUGCUUUAAAUUUAUUGGUUUGCACAGUUUAAGCGAUUUUCGUUUUGUAUUUUCCAUUUG-----CAGAC ----------------------.......-..........(((((((((((((...((((((....))))))..)))))))))))))......((((((.........))-----)))). ( -21.00) >DroSec_CAF1 10008 92 + 1 ----------------------UGUUUUG-UUUCGUGUUCAUCGCUUAAGCUGCUUUAAAUUUAUUGGUUUGCACAGUUUAAGCGAUUUUCGUUUUGUAUUUUCCAUUUG-----CAGAC ----------------------.......-..........(((((((((((((...((((((....))))))..)))))))))))))......((((((.........))-----)))). ( -21.00) >DroEre_CAF1 7396 92 + 1 ----------------------UGUUUUG-UUUCGUGUUCAUCGCUUAAGCUGCUUUAAAUUUAUUGGUUUGCACAGUUUAAGCGAUUUUCGUUUUGUAUUUUCCAUUUG-----CAGAC ----------------------.......-..........(((((((((((((...((((((....))))))..)))))))))))))......((((((.........))-----)))). ( -21.00) >DroAna_CAF1 5068 120 + 1 CGCCCCGGGCCAAGUGCAAUUUUUUAUUGUUUUUGCGCUCGUCGCUUAAGCUGCUUUAAAUUUAUUGGUUUGCACAGUUUAAGCGAUUUUCGUUUUGUAUUUUCCAUUUGGCCUGAAAAC .....(((((((((((((((.....)))).....(((...(((((((((((((...((((((....))))))..)))))))))))))...)))...........)))))))))))..... ( -39.20) >consensus ______________________UGUUUUG_UUUCGUGUUCAUCGCUUAAGCUGCUUUAAAUUUAUUGGUUUGCACAGUUUAAGCGAUUUUCGUUUUGUAUUUUCCAUUUG_____CAGAC ...................................((...(((((((((((((...((((((....))))))..)))))))))))))...))............................ (-19.05 = -18.67 + -0.37)


| Location | 20,099,771 – 20,099,863 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -13.90 |
| Energy contribution | -14.15 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20099771 92 - 22224390 GUCUG-----CAAAUGGAAAAUACAAAACGAAAAUCGCUUAAACUGUGCAAACCAAUAAAUUUAAAGCAGCUUAAGCGAUGAACACGAAA-CAAAACA---------------------- .(((.-----.....)))..........((...(((((((((.((((..(((........)))...)))).))))))))).....))...-.......---------------------- ( -15.20) >DroSec_CAF1 10008 92 - 1 GUCUG-----CAAAUGGAAAAUACAAAACGAAAAUCGCUUAAACUGUGCAAACCAAUAAAUUUAAAGCAGCUUAAGCGAUGAACACGAAA-CAAAACA---------------------- .(((.-----.....)))..........((...(((((((((.((((..(((........)))...)))).))))))))).....))...-.......---------------------- ( -15.20) >DroEre_CAF1 7396 92 - 1 GUCUG-----CAAAUGGAAAAUACAAAACGAAAAUCGCUUAAACUGUGCAAACCAAUAAAUUUAAAGCAGCUUAAGCGAUGAACACGAAA-CAAAACA---------------------- .(((.-----.....)))..........((...(((((((((.((((..(((........)))...)))).))))))))).....))...-.......---------------------- ( -15.20) >DroAna_CAF1 5068 120 - 1 GUUUUCAGGCCAAAUGGAAAAUACAAAACGAAAAUCGCUUAAACUGUGCAAACCAAUAAAUUUAAAGCAGCUUAAGCGACGAGCGCAAAAACAAUAAAAAAUUGCACUUGGCCCGGGGCG ........(((...(((.................((((((((.((((..(((........)))...)))).))))))))((((.((((.............)))).))))..))).))). ( -28.62) >consensus GUCUG_____CAAAUGGAAAAUACAAAACGAAAAUCGCUUAAACUGUGCAAACCAAUAAAUUUAAAGCAGCUUAAGCGAUGAACACGAAA_CAAAACA______________________ .............................(...(((((((((.((((..(((........)))...)))).)))))))))...).................................... (-13.90 = -14.15 + 0.25)



| Location | 20,099,788 – 20,099,903 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -18.62 |
| Energy contribution | -18.40 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20099788 115 + 22224390 AUCGCUUAAGCUGCUUUAAAUUUAUUGGUUUGCACAGUUUAAGCGAUUUUCGUUUUGUAUUUUCCAUUUG-----CAGACUCGCUGGUCAGCCCAAACAGCACACUAUUGCGUUUCUUCC (((((((((((((...((((((....))))))..)))))))))))))...(((((((((.........))-----))))...((((...........))))........)))........ ( -26.90) >DroEre_CAF1 7413 115 + 1 AUCGCUUAAGCUGCUUUAAAUUUAUUGGUUUGCACAGUUUAAGCGAUUUUCGUUUUGUAUUUUCCAUUUG-----CAGACUCGCUGGCCAGCCGAACCAGAGCACUACUGCUCCCUUUCC (((((((((((((...((((((....))))))..))))))))))))).((((.((((((.........))-----))))...(((....)))))))...(((((....)))))....... ( -30.70) >DroAna_CAF1 5108 100 + 1 GUCGCUUAAGCUGCUUUAAAUUUAUUGGUUUGCACAGUUUAAGCGAUUUUCGUUUUGUAUUUUCCAUUUGGCCUGAAAACCCACUCCCCAUCCCGGC-AGA------------------- (((((((((((((...((((((....))))))..))))))))))))).......................(((.....................)))-...------------------- ( -21.10) >consensus AUCGCUUAAGCUGCUUUAAAUUUAUUGGUUUGCACAGUUUAAGCGAUUUUCGUUUUGUAUUUUCCAUUUG_____CAGACUCGCUGGCCAGCCCAACCAGA_CACUA_UGC_____UUCC (((((((((((((...((((((....))))))..)))))))))))))......................................................................... (-18.62 = -18.40 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:44 2006