
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,098,382 – 20,098,528 |
| Length | 146 |
| Max. P | 0.999847 |

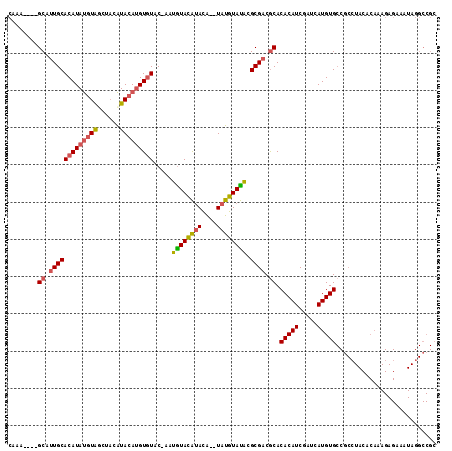
| Location | 20,098,382 – 20,098,491 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.20 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.55 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.69 |
| SVM decision value | 4.20 |
| SVM RNA-class probability | 0.999833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
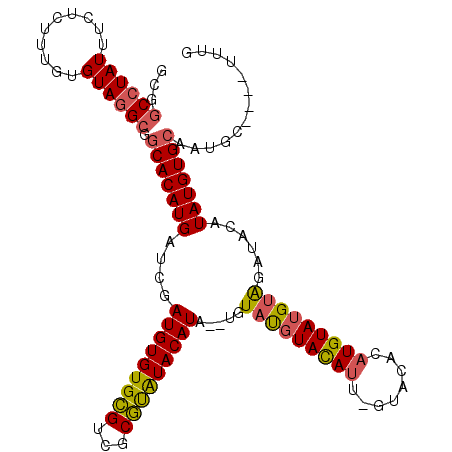
>X_DroMel_CAF1 20098382 109 + 22224390 CAAA----GCAUUGCACAUAUGUAUCCACAUACAUGUGUCC--AUAUACAUACAUGUAUGUAUACGCGACGCACACAUCGAUCAUGUGCCUCCUACACAAAGAGAAAUAGGCCGC ....----((...((((((((((((....))))))((((((--.((((((((....)))))))).).)))))...........))))))(((.........)))......))... ( -27.20) >DroSec_CAF1 8248 107 + 1 CA-A----GCAUUGCACAUAUGUAGCUCCAUACAUGUGUAC-AAUGUACACACA--UAUGUAUACGCGACGCACACAUCGAUCAUGUGCCGCAUACACAAAGAGAAAUAGGCCGC ..-.----((((((.(((((((((......))))))))).)-))))).......--..(((((..(((..(((((.........))))))))))))).................. ( -27.40) >DroSim_CAF1 8290 108 + 1 CAAA----GCAUUGCACAUAUGUAGCUACAUACAUGUGUAC-AAUGUACAUACA--UAUGUAUGCGCGACGCACACAUCGAUCAUGUGNNNNNNNNNNNNNNNNNNNNNNNNNNN ....----((((((.(((((((((......))))))))).)-)))))((((...--.((((.((((...)))).)))).....))))............................ ( -24.60) >DroEre_CAF1 6310 113 + 1 CAAAGCCGGCACUGCACAUACCUGUAUACGUACAAGUAUGCUUAUGUAUGUAUG--UAUGUACAUGCGCCACACACAUCGAUCAUGUGCCGCCUACACAAAGAGAAAUAGGCCGC ....((((((...((((((....((((((......))))))....((((((((.--...))))))))................)))))).))).........(....).)))... ( -29.90) >consensus CAAA____GCAUUGCACAUAUGUAGCUACAUACAUGUGUAC_AAUGUACAUACA__UAUGUAUACGCGACGCACACAUCGAUCAUGUGCCGCCUACACAAAGAGAAAUAGGCCGC ........((.(((((((((((((......))))))))).....((((((((....)))))))).)))).)).(((((.....)))))........................... (-18.92 = -19.55 + 0.63)


| Location | 20,098,382 – 20,098,491 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.20 |
| Mean single sequence MFE | -35.85 |
| Consensus MFE | -20.34 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.57 |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20098382 109 - 22224390 GCGGCCUAUUUCUCUUUGUGUAGGAGGCACAUGAUCGAUGUGUGCGUCGCGUAUACAUACAUGUAUGUAUAU--GGACACAUGUAUGUGGAUACAUAUGUGCAAUGC----UUUG (((((......(((((.....)))))((((((.......)))))))))))(((((((((....)))))))))--(.(((.((((((....)))))).))).).....----.... ( -35.90) >DroSec_CAF1 8248 107 - 1 GCGGCCUAUUUCUCUUUGUGUAUGCGGCACAUGAUCGAUGUGUGCGUCGCGUAUACAUA--UGUGUGUACAUU-GUACACAUGUAUGGAGCUACAUAUGUGCAAUGC----U-UG (((.((...........(((((((((((((((.......))))))..)))))))))(((--(((((((((...-)))))))))))))).((.((....))))..)))----.-.. ( -36.40) >DroSim_CAF1 8290 108 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNCACAUGAUCGAUGUGUGCGUCGCGCAUACAUA--UGUAUGUACAUU-GUACACAUGUAUGUAGCUACAUAUGUGCAAUGC----UUUG ............................(((((....(((((((((...))))))))).--..))))).((((-(((((.(((((......))))).))))))))).----.... ( -29.80) >DroEre_CAF1 6310 113 - 1 GCGGCCUAUUUCUCUUUGUGUAGGCGGCACAUGAUCGAUGUGUGUGGCGCAUGUACAUA--CAUACAUACAUAAGCAUACUUGUACGUAUACAGGUAUGUGCAGUGCCGGCUUUG ((.((((((..(.....).))))))(((((........((((((((....((((....)--))).)))))))).(((((((((((....)))))))))))...))))).)).... ( -41.30) >consensus GCGGCCUAUUUCUCUUUGUGUAGGCGGCACAUGAUCGAUGUGUGCGUCGCGUAUACAUA__UGUAUGUACAUU_GUACACAUGUAUGUAGAUACAUAUGUGCAAUGC____UUUG ...((((((..........)))))).(((((((....(((((((((...))))))))).....(((((((((........)))))))))......)))))))............. (-20.34 = -20.90 + 0.56)


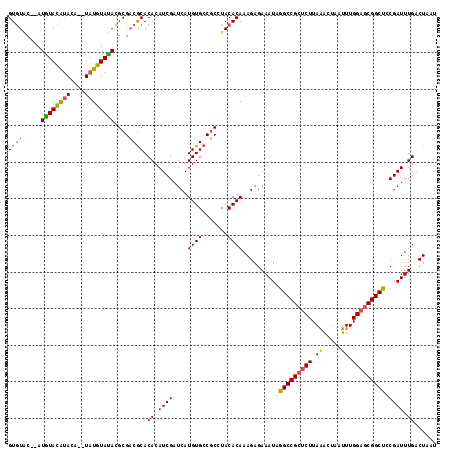
| Location | 20,098,413 – 20,098,528 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -25.68 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20098413 115 + 22224390 GUGUCC--AUAUACAUACAUGUAUGUAUACGCGACGCACACAUCGAUCAUGUGCCUCCUACACAAAGAGAAAUAGGCCGCUCUUAAACUAAUUUGGAACGGCUUCGAUUUGACUAAU ((((((--.((((((((....)))))))).).))))).((.(((((...((((.......))))..........(((((.(((...........))).)))))))))).))...... ( -31.80) >DroSec_CAF1 8278 114 + 1 GUGUAC-AAUGUACACACA--UAUGUAUACGCGACGCACACAUCGAUCAUGUGCCGCAUACACAAAGAGAAAUAGGCCGCUCUUAAACUAAUUUGGAGCGGCUCCGAUUUGACUAAU ((((((-(((((....)))--).)))))))(((..(((((.........)))))))).....((((..(.....(((((((((...........))))))))).)..))))...... ( -34.00) >DroEre_CAF1 6345 115 + 1 GUAUGCUUAUGUAUGUAUG--UAUGUACAUGCGCCACACACAUCGAUCAUGUGCCGCCUACACAAAGAGAAAUAGGCCGCACUUAGGCUAAUUUGGAGCGGCCCCGAUUUGCCUAAU ((.((....(((((((((.--...)))))))))...)).)).........((((.(((((............))))).))))((((((.((((.((......)).)))).)))))). ( -32.70) >consensus GUGUAC__AUGUACAUACA__UAUGUAUACGCGACGCACACAUCGAUCAUGUGCCGCCUACACAAAGAGAAAUAGGCCGCUCUUAAACUAAUUUGGAGCGGCUCCGAUUUGACUAAU ((((.....((((((((....))))))))....)))).((.((((....((((.......))))..........(((((((((.((.....)).))))))))).)))).))...... (-25.68 = -25.80 + 0.12)


| Location | 20,098,413 – 20,098,528 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -27.70 |
| Energy contribution | -27.72 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20098413 115 - 22224390 AUUAGUCAAAUCGAAGCCGUUCCAAAUUAGUUUAAGAGCGGCCUAUUUCUCUUUGUGUAGGAGGCACAUGAUCGAUGUGUGCGUCGCGUAUACAUACAUGUAUGUAUAU--GGACAC ...............(((((((..((.....))..)))))))......(((((.....)))))((((((.......))))))(((.((((((((((....)))))))))--)))).. ( -36.80) >DroSec_CAF1 8278 114 - 1 AUUAGUCAAAUCGGAGCCGCUCCAAAUUAGUUUAAGAGCGGCCUAUUUCUCUUUGUGUAUGCGGCACAUGAUCGAUGUGUGCGUCGCGUAUACAUA--UGUGUGUACAUU-GUACAC ...((..((((.((.(((((((..((.....))..)))))))))))))..)).((((((((((((((((.......))))))..))))))))))..--...((((((...-)))))) ( -38.40) >DroEre_CAF1 6345 115 - 1 AUUAGGCAAAUCGGGGCCGCUCCAAAUUAGCCUAAGUGCGGCCUAUUUCUCUUUGUGUAGGCGGCACAUGAUCGAUGUGUGUGGCGCAUGUACAUA--CAUACAUACAUAAGCAUAC .((((((.(((..(((....)))..))).))))))((((.((((((..(.....).)))))).))))(((....((((((((.....((((....)--)))))))))))...))).. ( -38.10) >consensus AUUAGUCAAAUCGGAGCCGCUCCAAAUUAGUUUAAGAGCGGCCUAUUUCUCUUUGUGUAGGCGGCACAUGAUCGAUGUGUGCGUCGCGUAUACAUA__UGUAUGUACAU__GCACAC .........(((((.(((((((.............))))))).......((..(((((.....))))).)))))))((((((.....(((((((((....)))))))))..)))))) (-27.70 = -27.72 + 0.02)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:41 2006