
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,096,859 – 20,097,000 |
| Length | 141 |
| Max. P | 0.985616 |

| Location | 20,096,859 – 20,096,960 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.09 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -14.14 |
| Energy contribution | -15.18 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20096859 101 + 22224390 UGUGAA-------GAUGCCCUAAAAUGCCUUUGUCAAUGCCAUGUC--AGGGAUAUUGGCUCCUAAUAUGGGGUAUCCUCGCUGGCA-GGAGACCCAAUA------AUGGCCCAC---CC .((((.-------(((((((((....(((..((((..((......)--)..))))..)))........))))))))).)))).(((.-(....).((...------.)))))...---.. ( -31.00) >DroSec_CAF1 6769 101 + 1 CGUGAG-------AAGGACCUAAAAUGCCUUUGUCAGUGCCAUGUC--AGGGAUAUUGGCCCCUAAUAAGGGGUAUCCUCGCUGGCA-GGAUACCCAGUC------GUGGCCCAC---CC .(((.(-------((((..........)))))......((((((.(--((((........))))......(((((((((.(....))-)))))))).).)------))))).)))---.. ( -36.40) >DroSim_CAF1 6792 100 + 1 CGUGAG-------AAGGACCUAAAAUGCCUUUGUCAGUGCCAUGUC--AGGGAUAUUGGCCCCUAAUAAGGGGUAUCCUCGCUGGCA-GGAUACCCAGUC------GUGGC-CAC---CC .(((.(-------((((..........)))))......((((((.(--((((........))))......(((((((((.(....))-)))))))).).)------)))))-)))---.. ( -36.40) >DroEre_CAF1 3698 101 + 1 CGUGAG-------GAGGAGCUAGAAUGUCUUUGUCAGUGCCAUGUC--AGGGAUUUUGGUCCCUGGCAUG-AGUGUCCUCGAUGGCAAGGACACCCAGGC------GUGGCCCAC---CC .(((.(-------(....(((.(..((((((((((((..(((((((--((((((....))))))))))))-.)..)......)))))))))))..).)))------....)))))---.. ( -45.90) >DroAna_CAF1 960 112 + 1 UGGGAAGCUGACUGAGAAACUAAAAUGUCUUUGUCAGGGAUAUUUUGGAGGGAGGAUGGGU----GGAUGGGCUAU---GGCUACCU-ACCUACCCAAUAAAAAGGAUCGCCCACACUCC ((((...(((((.((((..........)))).))))).(((.(((((..((((((.(((((----((.........---..))))))-)))).)))..)))))...))).))))...... ( -34.70) >consensus CGUGAG_______GAGGACCUAAAAUGCCUUUGUCAGUGCCAUGUC__AGGGAUAUUGGCCCCUAAUAUGGGGUAUCCUCGCUGGCA_GGAUACCCAGUC______GUGGCCCAC___CC ..........................(((...........(((((...((((........)))).)))))((((((((..........))))))))............)))......... (-14.14 = -15.18 + 1.04)



| Location | 20,096,892 – 20,097,000 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.72 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -19.14 |
| Energy contribution | -20.94 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20096892 108 + 22224390 CAUGUC--AGGGAUAUUGGCUCCUAAUAUGGGGUAUCCUCGCUGGCA-GGAGACCCAAUA------AUGGCCCAC---CCACGGCCAAUGGCCAUUCCAUUUCGCUCCCGGAGAAGUUUA ..((((--((.((.....(((((......)))))....)).))))))-((((........------.(((((...---....)))))((((.....))))....))))............ ( -32.30) >DroSec_CAF1 6802 108 + 1 CAUGUC--AGGGAUAUUGGCCCCUAAUAAGGGGUAUCCUCGCUGGCA-GGAUACCCAGUC------GUGGCCCAC---CCAUGGCCAAUGGCCAUUCCAUUUCGCUCCCGUAGAAGUUUA ......--..(((...(((((.........(((((((((.(....))-)))))))).(((------((((.....---)))))))....))))).)))(((((((....)).)))))... ( -38.20) >DroSim_CAF1 6825 107 + 1 CAUGUC--AGGGAUAUUGGCCCCUAAUAAGGGGUAUCCUCGCUGGCA-GGAUACCCAGUC------GUGGC-CAC---CCACGGCCAAUGGCCAUUCCAUUUCGCUGCCGUAGAAGUUUA .(((.(--((.((...(((((.........(((((((((.(....))-)))))))).(((------((((.-...---)))))))....))))).......)).))).)))......... ( -40.90) >DroEre_CAF1 3731 107 + 1 CAUGUC--AGGGAUUUUGGUCCCUGGCAUG-AGUGUCCUCGAUGGCAAGGACACCCAGGC------GUGGCCCAC---CCACGGCCAACGGCUAUUUCAUCUCGUU-CCGGAGAAGUUUA ((((((--((((((....))))))))))))-.(((((((........)))))))..((.(------((((((...---....)))).))).))......((((...-...))))...... ( -46.20) >DroAna_CAF1 1000 107 + 1 UAUUUUGGAGGGAGGAUGGGU----GGAUGGGCUAU---GGCUACCU-ACCUACCCAAUAAAAAGGAUCGCCCACACUCCACGGGCCACGC-----CCACUUCUCUCUCAACAAAGUUUA .((((((((((((((((((((----(..((((.((.---((......-.))))))))............((((.........))))..)))-----))).)))))))))..))))))... ( -37.40) >consensus CAUGUC__AGGGAUAUUGGCCCCUAAUAUGGGGUAUCCUCGCUGGCA_GGAUACCCAGUC______GUGGCCCAC___CCACGGCCAAUGGCCAUUCCAUUUCGCUCCCGGAGAAGUUUA ..........(((...(((((.........((((((((..........))))))))...........(((((..........)))))..))))).)))...................... (-19.14 = -20.94 + 1.80)

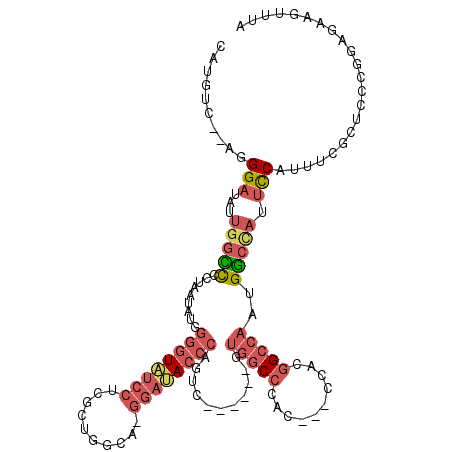
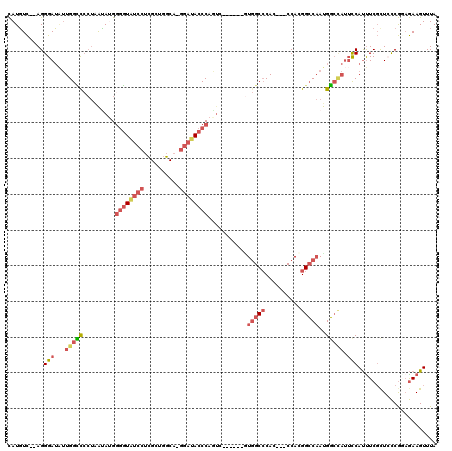
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:36 2006