
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,072,769 – 20,072,913 |
| Length | 144 |
| Max. P | 0.729063 |

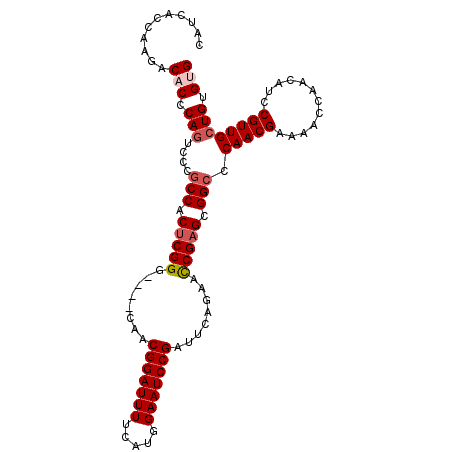
| Location | 20,072,769 – 20,072,874 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -24.23 |
| Energy contribution | -25.07 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
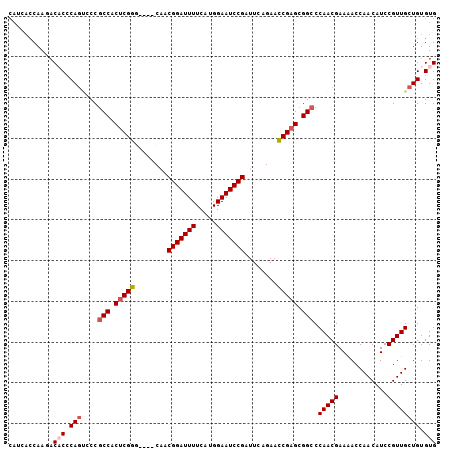
>X_DroMel_CAF1 20072769 105 + 22224390 CAUCACCAAGACACCCAGUGCCGCCACUCGGGGCCACUACGGAUUUUCAUGGAAUCCGAUUCAGAACCGAGCGGCCCAACGAAAACCAACAUCCGUUGCUGUGUG ...........(((.(((....(((.(((((..(.....(((((((.....))))))).....)..))))).))).(((((............)))))))).))) ( -32.00) >DroSec_CAF1 9571 101 + 1 CAUCACCAAGACACCCAGUCCCGCCGCUCGGG----CAACGGAUUUUCAUGGAAUCCGAUUCAGAACCGAGCGGCCCAACGAAAACCAACAUCCGUUGCUGUGUG ...........(((.(((....(((((((((.----...(((((((.....)))))))........))))))))).(((((............)))))))).))) ( -34.40) >DroSim_CAF1 23799 101 + 1 CAUCACCAAGACACCCAGUCCCGCCGCUCGGG----CAACGGAUUUUCAUGGAAUCCGAUUCGGAACCGAGCGGCCCAACGAAAACCAACAUCCGUUGCUGUGUG ...........(((.(((....(((((((((.----...(((((((.....)))))))........))))))))).(((((............)))))))).))) ( -34.40) >DroEre_CAF1 8701 101 + 1 CAGAACCAACACCCCCAGCCCCACCACUCGGA----CACCGGAUUUUCAUGGAAUCCGAUUCGGAAUCGAGCGGCCCAACGAAGACCAACAUCCGUUGCUGUGUG ........................(((.(((.----((.(((((((((.(((...(((.((((....))))))).)))..)))))......)))).))))).))) ( -22.00) >DroYak_CAF1 9797 95 + 1 ------CAAACCCCACAAACCCGCCACACGGA----CUCCGGAUUUUCAUGGAAUCCGAUUCAGAACCGAGCGGCCCAACGAAAACCAACAUCCGUUGCUGUGUG ------..................(((((((.----(..(((((((((.(((...(((.(((......)))))).)))..))))......)))))..)))))))) ( -23.10) >consensus CAUCACCAAGACACCCAGUCCCGCCACUCGGG____CAACGGAUUUUCAUGGAAUCCGAUUCAGAACCGAGCGGCCCAACGAAAACCAACAUCCGUUGCUGUGUG ...........(((.(((....(((.(((((........(((((((.....)))))))........))))).))).(((((............)))))))).))) (-24.23 = -25.07 + 0.84)

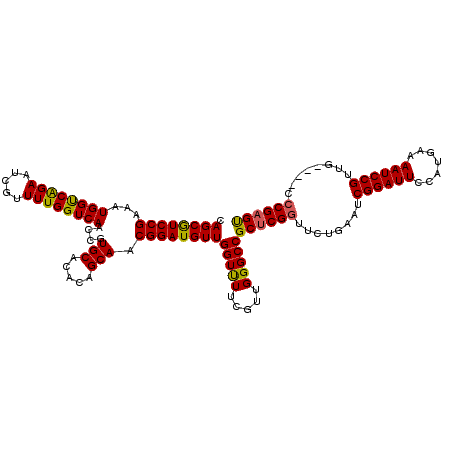
| Location | 20,072,794 – 20,072,913 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -40.94 |
| Consensus MFE | -35.57 |
| Energy contribution | -35.01 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
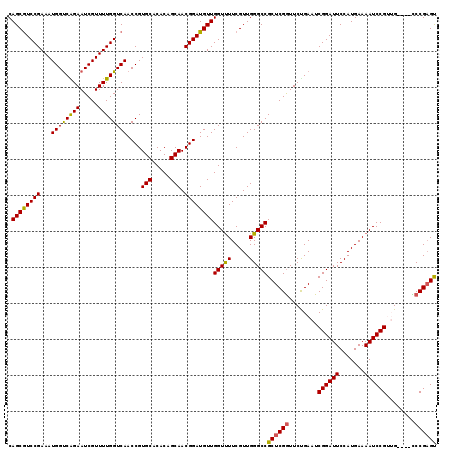
>X_DroMel_CAF1 20072794 119 - 22224390 CAGCGUCCGAAAUGGUCAGAAUCGUUUUGGUCAACCGUGCACACAGCAACGGAUGUUGGUUUUCGUUGGGCCGCUCGGUUCUGAAUCGGAUUCCAUGAAAAUCCGUAGUGGCCCCGAGU (((((((((...((..((((.....))))..))....(((.....))).)))))))))...((((..((((((((((((((.((((...))))...)))...))).)))))))))))). ( -44.10) >DroSec_CAF1 9596 115 - 1 CAGCGUCCGAAAUGGUCGGAAUCGUUUUGGUCAACCGUGCACACAGCAACGGAUGUUGGUUUUCGUUGGGCCGCUCGGUUCUGAAUCGGAUUCCAUGAAAAUCCGUUG----CCCGAGC ..(((.((((((((((....)))))))))).).............(((((((((..(((..((((..(((((....))))).....))))..))).....))))))))----)....)) ( -39.70) >DroSim_CAF1 23824 115 - 1 CAGCGUCCGAAAUGGUCGGAAUCGUUUUGGUCAACCGUGCACACAGCAACGGAUGUUGGUUUUCGUUGGGCCGCUCGGUUCCGAAUCGGAUUCCAUGAAAAUCCGUUG----CCCGAGC ..(((.((((((((((....)))))))))).).............(((((((((..(((..(((((((((((....)).)))))..))))..))).....))))))))----)....)) ( -40.40) >DroEre_CAF1 8726 115 - 1 CAGCGUCCGAAAUGGUCAGAAUCGUUUUGGUCAACCGUGCACACAGCAACGGAUGUUGGUCUUCGUUGGGCCGCUCGAUUCCGAAUCGGAUUCCAUGAAAAUCCGGUG----UCCGAGU .((((((((...((..((((.....))))..))....(((.....))).))))))))(((((.....)))))(((((.......((((((((.......)))))))).----..))))) ( -37.60) >DroYak_CAF1 9816 115 - 1 CAGCAUCCGAAAUGGCCAGAAUCGUUUUGGUCAACCGUGCACACAGCAACGGAUGUUGGUUUUCGUUGGGCCGCUCGGUUCUGAAUCGGAUUCCAUGAAAAUCCGGAG----UCCGUGU (((((((((...((((((((.....))))))))....(((.....))).)))))))))...((((..(((((....))))))))).((((((((((....))..))))----))))... ( -42.90) >consensus CAGCGUCCGAAAUGGUCAGAAUCGUUUUGGUCAACCGUGCACACAGCAACGGAUGUUGGUUUUCGUUGGGCCGCUCGGUUCUGAAUCGGAUUCCAUGAAAAUCCGUUG____CCCGAGU .((((((((...((((((((.....))))))))....(((.....))).))))))))(((((.....)))))((((((........((((((.......))))))........)))))) (-35.57 = -35.01 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:20 2006