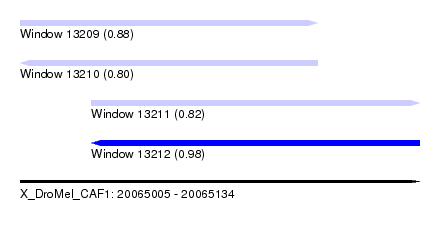
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,065,005 – 20,065,134 |
| Length | 129 |
| Max. P | 0.980606 |

| Location | 20,065,005 – 20,065,101 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 88.91 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -22.62 |
| Energy contribution | -22.62 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20065005 96 + 22224390 CGCAUCGUUUUUAUGUGCAUUCGCGCCCGCACACCCAAUUUAUAUAUAAUAUAUAUGUAUGUAUCUGUGUACACUUAAGGGGGCUUGGGUGUGCGC ..............((((....)))).(((((((((((..((((((((...))))))))(((((....)))))...........))))))))))). ( -28.60) >DroSim_CAF1 1177 93 + 1 CGCAUCGUUUUUAUGUGCAUUCGCGCCCGCACACCCAAUUUAUAUAU---AUAUAUACAUGUAUCUGUGUACACUUAAGGGGGCGUGGGUGUGCGC .(((.(((....))))))...(((((((((.(.(((........(((---(((.(((....))).))))))........)))).)))))))))... ( -29.29) >DroEre_CAF1 2596 83 + 1 CGCAUCGUUUUUAUGUGCAUUCGCGCUCGCACACCCAAUUUAUAUAUA-------------UAUCUGUGUACACUUAAGGGGGCGUGGGUGUGCGC .(((.(((....))))))...(((((.(.(((.(((..((..((((((-------------....)))))).....))..))).))).).))))). ( -24.90) >DroYak_CAF1 2665 83 + 1 CGCAUCGUUUUUAUGUGCAUUCGCGCUCGCACACCCAAUUUAUAUAUA-------------UAUCUGUGUACACUUAAGGGGGCGUGGGUGUGCGC .(((.(((....))))))...(((((.(.(((.(((..((..((((((-------------....)))))).....))..))).))).).))))). ( -24.90) >consensus CGCAUCGUUUUUAUGUGCAUUCGCGCCCGCACACCCAAUUUAUAUAUA_____________UAUCUGUGUACACUUAAGGGGGCGUGGGUGUGCGC ..............(..((((((((((((((((................................)))))...(.....))))))))))))..).. (-22.62 = -22.62 + -0.00)



| Location | 20,065,005 – 20,065,101 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 88.91 |
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -18.73 |
| Energy contribution | -19.23 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20065005 96 - 22224390 GCGCACACCCAAGCCCCCUUAAGUGUACACAGAUACAUACAUAUAUAUUAUAUAUAAAUUGGGUGUGCGGGCGCGAAUGCACAUAAAAACGAUGCG .(((((((((((..........(((((......)))))...((((((...))))))..))))))))))).(((((..............)).))). ( -26.04) >DroSim_CAF1 1177 93 - 1 GCGCACACCCACGCCCCCUUAAGUGUACACAGAUACAUGUAUAUAU---AUAUAUAAAUUGGGUGUGCGGGCGCGAAUGCACAUAAAAACGAUGCG ((((...(((((((((......((((......)))).((((((...---.))))))....))))))).))))))....((((........).))). ( -26.10) >DroEre_CAF1 2596 83 - 1 GCGCACACCCACGCCCCCUUAAGUGUACACAGAUA-------------UAUAUAUAAAUUGGGUGUGCGAGCGCGAAUGCACAUAAAAACGAUGCG .(((((((((((((........)))).......((-------------(....)))....))))))))).(((((..............)).))). ( -21.34) >DroYak_CAF1 2665 83 - 1 GCGCACACCCACGCCCCCUUAAGUGUACACAGAUA-------------UAUAUAUAAAUUGGGUGUGCGAGCGCGAAUGCACAUAAAAACGAUGCG .(((((((((((((........)))).......((-------------(....)))....))))))))).(((((..............)).))). ( -21.34) >consensus GCGCACACCCACGCCCCCUUAAGUGUACACAGAUA_____________UAUAUAUAAAUUGGGUGUGCGAGCGCGAAUGCACAUAAAAACGAUGCG ((((...(.(((((((..(((.(((((.....................))))).)))...))))))).).))))....((((........).))). (-18.73 = -19.23 + 0.50)


| Location | 20,065,028 – 20,065,134 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 84.84 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -26.03 |
| Energy contribution | -25.79 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20065028 106 + 22224390 CGCCCGCACACCCAAUUUAUAUAUAAUAUAUAUGUAUGUAUCUGUGUACACUUAAGGGGGCUUGGGUGUGCGCGCGUAUAUAUCUCCAUGCUUGUAUGUAAGGAC-------G (((.(((((((((((..((((((((...))))))))(((((....)))))...........))))))))))).)))........(((.(((......))).))).-------. ( -34.60) >DroSim_CAF1 1200 103 + 1 CGCCCGCACACCCAAUUUAUAUAU---AUAUAUACAUGUAUCUGUGUACACUUAAGGGGGCGUGGGUGUGCGCGCGUGUAUAUCUCCAUGCUUGUAUGUAAGGAC-------G (((.((((((((((...((((((.---(((((....))))).))))))(.(((....))).))))))))))).)))........(((.(((......))).))).-------. ( -32.90) >DroEre_CAF1 2619 93 + 1 CGCUCGCACACCCAAUUUAUAUAUA-------------UAUCUGUGUACACUUAAGGGGGCGUGGGUGUGCGCGCGUGUAUAUCUCCAUGUUUCUAUGUAAGGAC-------G (((.((((((((((...((((((..-------------....))))))(.(((....))).))))))))))).)))........(((.((........)).))).-------. ( -26.20) >DroYak_CAF1 2688 100 + 1 CGCUCGCACACCCAAUUUAUAUAUA-------------UAUCUGUGUACACUUAAGGGGGCGUGGGUGUGCGCGCGUGUAUAUCUCCAUGUUUCUAUGUAAGGAUCGUGUAAG (((.((((((((((...((((((..-------------....))))))(.(((....))).))))))))))).)))..........((((.((((.....)))).)))).... ( -30.30) >consensus CGCCCGCACACCCAAUUUAUAUAUA_____________UAUCUGUGUACACUUAAGGGGGCGUGGGUGUGCGCGCGUGUAUAUCUCCAUGCUUCUAUGUAAGGAC_______G (((.((((((((((...((((((...................))))))..((....))....)))))))))).)))........(((.(((......))).)))......... (-26.03 = -25.79 + -0.25)


| Location | 20,065,028 – 20,065,134 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 84.84 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -23.74 |
| Energy contribution | -23.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20065028 106 - 22224390 C-------GUCCUUACAUACAAGCAUGGAGAUAUAUACGCGCGCACACCCAAGCCCCCUUAAGUGUACACAGAUACAUACAUAUAUAUUAUAUAUAAAUUGGGUGUGCGGGCG .-------.(((..............)))........(((.(((((((((((..........(((((......)))))...((((((...))))))..))))))))))).))) ( -32.04) >DroSim_CAF1 1200 103 - 1 C-------GUCCUUACAUACAAGCAUGGAGAUAUACACGCGCGCACACCCACGCCCCCUUAAGUGUACACAGAUACAUGUAUAUAU---AUAUAUAAAUUGGGUGUGCGGGCG .-------.(((..............)))........(((.((((((((((........((.(((((((........))))))).)---).........)))))))))).))) ( -31.67) >DroEre_CAF1 2619 93 - 1 C-------GUCCUUACAUAGAAACAUGGAGAUAUACACGCGCGCACACCCACGCCCCCUUAAGUGUACACAGAUA-------------UAUAUAUAAAUUGGGUGUGCGAGCG .-------.(((..............)))........(((.(((((((((((((........)))).......((-------------(....)))....))))))))).))) ( -25.74) >DroYak_CAF1 2688 100 - 1 CUUACACGAUCCUUACAUAGAAACAUGGAGAUAUACACGCGCGCACACCCACGCCCCCUUAAGUGUACACAGAUA-------------UAUAUAUAAAUUGGGUGUGCGAGCG .........(((..............)))........(((.(((((((((((((........)))).......((-------------(....)))....))))))))).))) ( -25.84) >consensus C_______GUCCUUACAUACAAACAUGGAGAUAUACACGCGCGCACACCCACGCCCCCUUAAGUGUACACAGAUA_____________UAUAUAUAAAUUGGGUGUGCGAGCG .........(((..............)))........(((.((((((((((.((........))(....).............................)))))))))).))) (-23.74 = -23.74 + 0.00)

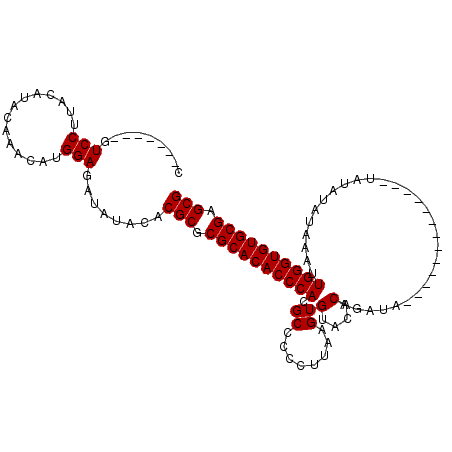

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:13 2006