
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,047,020 – 20,047,211 |
| Length | 191 |
| Max. P | 0.988167 |

| Location | 20,047,020 – 20,047,138 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -19.89 |
| Energy contribution | -22.07 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20047020 118 + 22224390 UCUUAGCGGGAUUCCGAAUUCUUAAUUAACACAAGUUUAAACACAAGAAUAAGAUUUUACUACUGUACGGCGAUAGCUGUGAACAGCUGUCGUGACAGCGAUAGUUCCAGUCUUUCCA ........(((......((((((....(((....))).......))))))((((((..((((((((...((((((((((....)))))))))).))))...))))...))))))))). ( -31.90) >DroSim_CAF1 3789 118 + 1 CCCUAGCGGAAUUCCCAAUUCUCAAUUAACACAGGUUUAAACACAAGAAUAAGCUUUUACUGCUGUACGACGAUAGUUGUGAACAGCUGUCGUGACAGCGAUAGUUCCACUACUCCCA ...(((.((((((..(................(((((((..........))))))).....(((((...((((((((((....)))))))))).))))))..)))))).)))...... ( -33.10) >DroEre_CAF1 1519 111 + 1 CUUUAGCGGAAU-------ACCUAAUUUACACAGGUUUAGGCACACGAAUAAGCUUUUAUUGUUGUACAGCGUUAGCUGAGAACAGCUGUCGUGACUGCGAUAGUUACCCUACUCUCG .....((((...-------.(((((...........)))))..(((((...((((......(((.(.((((....))))).))))))).))))).))))((.(((......))).)). ( -29.00) >DroYak_CAF1 2508 111 + 1 CUUUAGCGGAAU-------UCUUAAUUAUUACAGUGUUAAACACAAGAAUUAGUUAUUAAUACUCUAUGACGAUAGCUGUGAACAGCUGUCGUGACAGCGAUAGUUCCACUGCUCCCA ...(((.(((((-------(.((.........((((((((..((........))..))))))))((.(.((((((((((....)))))))))).).)).)).)))))).)))...... ( -30.40) >consensus CCUUAGCGGAAU_______UCUUAAUUAACACAGGUUUAAACACAAGAAUAAGCUUUUACUACUGUACGACGAUAGCUGUGAACAGCUGUCGUGACAGCGAUAGUUCCACUACUCCCA ...(((.(((((.....................(((..((((..........))))..)))(((((...((((((((((....)))))))))).)))))....))))).)))...... (-19.89 = -22.07 + 2.19)

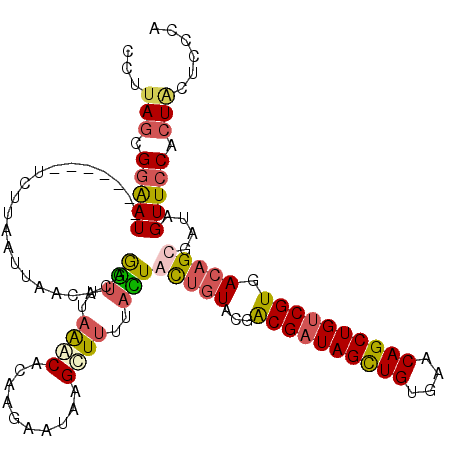

| Location | 20,047,059 – 20,047,178 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -38.05 |
| Consensus MFE | -24.61 |
| Energy contribution | -27.43 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20047059 119 + 22224390 AACACAAGAAUAAGAUUUUACUACUGUACGGCGAUAGCUGUGAACAGCUGUCGUGACAGCGAUAGUUCCAGUCUUUCCAUGCACUAGCGCAGCGAUCUGGCAGCACUACUGCGUUUAGC ...........((((((..((((((((...((((((((((....)))))))))).))))...))))...))))))...(((((.(((.((.((......)).)).))).)))))..... ( -38.90) >DroSim_CAF1 3828 119 + 1 AACACAAGAAUAAGCUUUUACUGCUGUACGACGAUAGUUGUGAACAGCUGUCGUGACAGCGAUAGUUCCACUACUCCCAUGCUCUAGCGCAGCGGUCUGGCAGCACGGCUGCGCUUAAC ............(((.......(((((...((((((((((....)))))))))).)))))(((((.....))).))....)))..((((((((.(((.....).)).)))))))).... ( -42.50) >DroEre_CAF1 1551 119 + 1 GGCACACGAAUAAGCUUUUAUUGUUGUACAGCGUUAGCUGAGAACAGCUGUCGUGACUGCGAUAGUUACCCUACUCUCGUUCUCUAGCGCAACGAUCUGGCAACACUGCUGCGGCUAGC (((...((....(((......((((((.(((((((.((((((((((((((((((....)))))))))...........)))))).)))..))))..)))))))))..))).)))))... ( -35.60) >DroYak_CAF1 2540 119 + 1 AACACAAGAAUUAGUUAUUAAUACUCUAUGACGAUAGCUGUGAACAGCUGUCGUGACAGCGAUAGUUCCACUGCUCCCAUGCUCUAGCGCAACGAUCUGGCAGCACUGCUGCGCUUAGC ..(((........(((((.........)))))((((((((....)))))))))))..((((.((((.....((((.((((((......)))......))).))))..)))))))).... ( -35.20) >consensus AACACAAGAAUAAGCUUUUACUACUGUACGACGAUAGCUGUGAACAGCUGUCGUGACAGCGAUAGUUCCACUACUCCCAUGCUCUAGCGCAACGAUCUGGCAGCACUGCUGCGCUUAGC .................(((.((((((...((((((((((....)))))))))).)))))).)))..................(((((((((((...((....)).))))))).)))). (-24.61 = -27.43 + 2.81)


| Location | 20,047,098 – 20,047,211 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 86.28 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -25.61 |
| Energy contribution | -27.30 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20047098 113 + 22224390 GUGAACAGCUGUCGUGACAGCGAUAGUUCCAGUCUUUCCAUGCACUAGCGCAGCGAUCUGGCAGCACUACUGCGUUUAGCAGAACGCUGUUAGUUACAGUUACAUUUCACAUA (((((.((((((..((((((((...(((((((....((..(((......)))..)).)))).)))....((((.....))))..))))))))...))))))....)))))... ( -38.20) >DroSim_CAF1 3867 113 + 1 GUGAACAGCUGUCGUGACAGCGAUAGUUCCACUACUCCCAUGCUCUAGCGCAGCGGUCUGGCAGCACGGCUGCGCUUAACAGCACGCUGUUAGUUACAGUUACAUUUCGCAUA (((((.((((((..((((((((.(((.....)))......((((..((((((((.(((.....).)).))))))))....))))))))))))...))))))....)))))... ( -42.90) >DroEre_CAF1 1590 113 + 1 GAGAACAGCUGUCGUGACUGCGAUAGUUACCCUACUCUCGUUCUCUAGCGCAACGAUCUGGCAACACUGCUGCGGCUAGCAGCACACUUUAAGUUACAGUUAUAUAUCGCUUA (((((((((((((((....)))))))))...........)))))).((((........((....)).((((((.....)))))).......................)))).. ( -30.60) >DroYak_CAF1 2579 113 + 1 GUGAACAGCUGUCGUGACAGCGAUAGUUCCACUGCUCCCAUGCUCUAGCGCAACGAUCUGGCAGCACUGCUGCGCUUAGCAGCACACUGUAAGUUACAGUUACAUUUCGCCUA (((((.(((((((((....)))))))))....((((.((((((......)))......))).)))).((((((.....)))))).((((((...)))))).....)))))... ( -34.30) >consensus GUGAACAGCUGUCGUGACAGCGAUAGUUCCACUACUCCCAUGCUCUAGCGCAACGAUCUGGCAGCACUGCUGCGCUUAGCAGCACACUGUAAGUUACAGUUACAUUUCGCAUA ((((..(((((((((....))))))))).....................(.(((...((((((((...(((((.....)))))..)))))))).....))).)...))))... (-25.61 = -27.30 + 1.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:03 2006