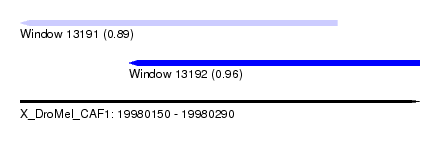
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,980,150 – 19,980,290 |
| Length | 140 |
| Max. P | 0.962321 |

| Location | 19,980,150 – 19,980,261 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.22 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
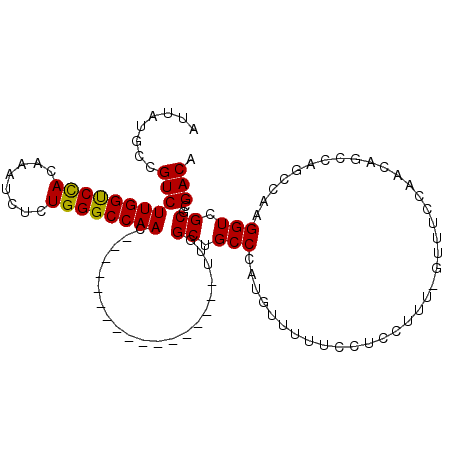
>X_DroMel_CAF1 19980150 111 - 22224390 AUUAUACCGUCUUGGCCUACAAAUCUCUGGGCCAACACUUAGCUUACCAUAUUUUAGCUGCCUAUAUUUUGUCUCCUUUCCUUUCCAACAGCCAGUCAAGGUCGCGCGACA ........((.((((((((........)))))))).)).(((((...........))))).........((((.(....((((..(........)..))))....).)))) ( -21.80) >DroSec_CAF1 18071 93 - 1 AUGAUGCCGUCUUGGUCCACAAAUCUCUGGGCCAAC-----------------UUGGCUGCCCAUGUUUUUCUUCCUUU-GUUUCCAACAGGCAGGCAAGGUCGCGCGACA .((.(((((.(((((((((........))))))))(-----------------(((.(((((..((((...........-......))))))))).))))).)).))).)) ( -30.03) >DroSim_CAF1 11092 93 - 1 AUUAUGCCGUCUUGGUCCACAAAUCUCUGGGCCAAC-----------------UUGGCUGCCCAUGUUUUUCCUCCUUU-GUUUCCAACAGCCAGCCAAGGUCGCGCGACA ....(((((.(((((((((........))))))))(-----------------(((((((....((((...........-......))))..))))))))).)).)))... ( -28.43) >consensus AUUAUGCCGUCUUGGUCCACAAAUCUCUGGGCCAAC_________________UUGGCUGCCCAUGUUUUUCCUCCUUU_GUUUCCAACAGCCAGCCAAGGUCGCGCGACA ........(((((((((((........)))))))).....................((.(((.....................................))).))..))). (-17.66 = -17.22 + -0.44)

| Location | 19,980,188 – 19,980,290 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.12 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.42 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19980188 102 - 22224390 UACGGGCACCUGUUCUU-UGGGCCAACUUAAUUAUACCGUCUUGGCCUACAAAUCUCUGGGCCAACACUUAGCUUACCAUAUUUUAGCUGC----CUAUAUUUUGUC .((((....))))....-(((((...............((.((((((((........)))))))).))..((((...........))))))----)))......... ( -24.00) >DroSec_CAF1 18108 85 - 1 CACUGGCACCUGUUCUU-UUGGCCAAUUUAAUGAUGCCGUCUUGGUCCACAAAUCUCUGGGCCAAC-----------------UUGGCUGC----CCAUGUUUUUCU ((..((((..((..(..-..)..))..........((((..((((((((........)))))))).-----------------.)))))))----)..))....... ( -23.80) >DroSim_CAF1 11129 85 - 1 CACUGGCACCUGUUCUU-UUGGCCAACUUAAUUAUGCCGUCUUGGUCCACAAAUCUCUGGGCCAAC-----------------UUGGCUGC----CCAUGUUUUUCC ((..((((..((..(..-..)..))..........((((..((((((((........)))))))).-----------------.)))))))----)..))....... ( -23.80) >DroEre_CAF1 13192 89 - 1 CAGUGGCACCUGUUCCU-UUGGCCAUGCUAAUUACGCCGUCUUGGUCCACAAAUCUCUGGGCCAAC-----------------CUAGCUGCUUAGCCAUGUUUUUCU ..(((((....((....-(((((...)))))..))((.((.((((((((........)))))))).-----------------...)).))...)))))........ ( -23.20) >DroYak_CAF1 10676 86 - 1 CAGUGGCACCUGUUCUUUUUGGCCAAUUUAAUUAUGCCGCCCUGGUCUACAAAUCUCUGGGCCAAC-----------------UUAGCU----CGCAAUAUAUAUUU ..(((((.............(((............)))((((.(((......)))...))))....-----------------...)).----)))........... ( -16.00) >consensus CACUGGCACCUGUUCUU_UUGGCCAACUUAAUUAUGCCGUCUUGGUCCACAAAUCUCUGGGCCAAC_________________UUAGCUGC____CCAUGUUUUUCU ....(((.............(((............)))...((((((((........)))))))).....................))).................. (-14.42 = -14.42 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:50 2006