
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,978,658 – 19,978,809 |
| Length | 151 |
| Max. P | 0.791779 |

| Location | 19,978,658 – 19,978,778 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -15.69 |
| Energy contribution | -15.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19978658 120 - 22224390 UGGAUAACAGGACAGUCUGGUCCUUCCACAACAAACGCCAUCAUGAUGGCUCAUUAUAUUCAUCGACUGUUUAAUCGACCCCAUCAUACCAACCCCAACUAUCCACAAGCACGAACGCAA ((((((...(((((((((((.....)))........(((((....)))))..............))))))))....((.....))..............))))))...((......)).. ( -24.90) >DroSec_CAF1 17253 120 - 1 UGGAUAACAGGACAGCCUGGUUCAUCUACAACAAACGCCAUUAUAAUGGCUCCUUACAUUCAUCGACUGCUUAAUCGACCCCUUCAUACCAACCCCAAUUAUUCACAAGCACGAACACAA .......((((....))))((((.............(((((....)))))............((((........))))..................................)))).... ( -17.70) >DroSim_CAF1 10289 113 - 1 UGGACAACAGGACAGCCUGGUUCAUCUACAACAAACGCCAUUAUAAUGGCUCCUUACAUUCAUCGACUGCUUAAUCGACCCCUUCAUACCAACCCCAACU-------AGCACGAACACAA .......((((....))))((((.............(((((....)))))............((((........))))......................-------.....)))).... ( -17.70) >consensus UGGAUAACAGGACAGCCUGGUUCAUCUACAACAAACGCCAUUAUAAUGGCUCCUUACAUUCAUCGACUGCUUAAUCGACCCCUUCAUACCAACCCCAACUAU_CACAAGCACGAACACAA .......((((....))))((((.............(((((....)))))............((((........))))..................................)))).... (-15.69 = -15.80 + 0.11)


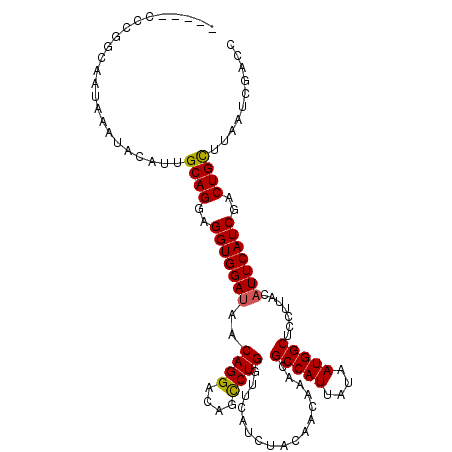
| Location | 19,978,698 – 19,978,809 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.02 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -23.87 |
| Energy contribution | -23.77 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19978698 111 - 22224390 AAAGUCCAGGCAAUAAAGACAUUGCAGAAGGUGGAUAACAGGACAGUCUGGUCCUUCCACAACAAACGCCAUCAUGAUGGCUCAUUAUAUUCAUCGACUGUUUAAUCGACC ..((((...(((((......))))).(((.(((((....(((((......)))))))))).......(((((....)))))........)))...))))............ ( -29.10) >DroSec_CAF1 17293 106 - 1 -----CCCGGCAAUAAAUACAUUGCAGGAGGUGGAUAACAGGACAGCCUGGUUCAUCUACAACAAACGCCAUUAUAAUGGCUCCUUACAUUCAUCGACUGCUUAAUCGACC -----....(((((......)))))((((.((((((..((((....))))....)))))).......(((((....)))))))))........((((........)))).. ( -26.80) >DroSim_CAF1 10322 106 - 1 -----CCCGGCAAUAAAUACAUUGCAGGAGGUGGACAACAGGACAGCCUGGUUCAUCUACAACAAACGCCAUUAUAAUGGCUCCUUACAUUCAUCGACUGCUUAAUCGACC -----.((.(((((......))))).))((((((((..((((....)))))))))))).........(((((....)))))............((((........)))).. ( -27.90) >consensus _____CCCGGCAAUAAAUACAUUGCAGGAGGUGGAUAACAGGACAGCCUGGUUCAUCUACAACAAACGCCAUUAUAAUGGCUCCUUACAUUCAUCGACUGCUUAAUCGACC .......................((((..(((((((..((((....)))).................(((((....))))).......)))))))..)))).......... (-23.87 = -23.77 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:49 2006