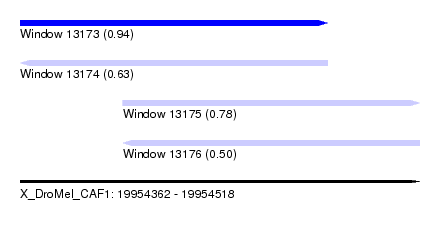
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,954,362 – 19,954,518 |
| Length | 156 |
| Max. P | 0.936547 |

| Location | 19,954,362 – 19,954,482 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.92 |
| Mean single sequence MFE | -34.76 |
| Consensus MFE | -25.14 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19954362 120 + 22224390 UAAAUUUAAAUGGCAAGGUGGCAGAACAUUUGAUCACAUUGCACUUGAUUGAAUUCCCGCUGAGGGAAUCAACCUCUUUUGAGUAACACCAAAUUUCAUAUGGGAAUUUGAUUCCAUUGA ........((((((((.((((((((...)))).)))).))))....(((..((((((((..((((.......)))).((((........)))).......))))))))..))).)))).. ( -30.60) >DroSec_CAF1 66862 120 + 1 CUUAUUUAAAUGGCAAGGUGGCGUUACAUUUGAUCACAUUAGACUUGAUUGAAUUCCCGCCGAGGGAAUCAAUCUCUUUCGAGAAACACCAAAUUCCAUAUGGGAAUUUGAUUCCGUCGA ................(((((.(....(((..((((.........))))..))).))))))((.((((((..((((....))))......(((((((.....))))))))))))).)).. ( -34.60) >DroSim_CAF1 56244 120 + 1 CUUAUUUAAAUGGCAAGGUGGCGUUACAUUUGAUCACAUUAGACUUGAUUGAAUUCCCGCCGAGGGAAUCAAUCUCUUUCGAGAAACACCAAAUUCCAUAUGGGAAUUUGAUUCCAUCGA ................(((((.(....(((..((((.........))))..))).))))))((.((((((..((((....))))......(((((((.....))))))))))))).)).. ( -32.50) >DroEre_CAF1 53619 120 + 1 CUUAUUUAAUUGACAAGGUGGCGGGAAACUCGAUCACAUUAGACCUGAUAGAAUUCCCGCUGUGGGAAUCAAUCUCUUUUGCGAAACACAAAAUUCCAUAUGGGAAUUUGAUUCCGUCGA .........(((((...(..(((((((..((.((((.........)))).)).)))))))..).((((((((((((...((.(((........)))))...))))..))))))))))))) ( -36.20) >DroYak_CAF1 55969 120 + 1 CUUAUUUAAGUGGCGAGGUGGCGGGACAUUUGAUCACAUUAGACUUGAUUGAAUUCCCGCUGUGGGAAUCAAUCUCUUUGGAGAAACACAGAAUUCCAUAUGGGAAUUUGAUUCGGUCGA .........((((..(((((......)))))..))))....((((.(((..((((((((.(((((((.((..((((....))))......)).)))))))))))))))..))).)))).. ( -39.90) >consensus CUUAUUUAAAUGGCAAGGUGGCGGGACAUUUGAUCACAUUAGACUUGAUUGAAUUCCCGCUGAGGGAAUCAAUCUCUUUCGAGAAACACCAAAUUCCAUAUGGGAAUUUGAUUCCGUCGA ..........((((.....(((((...(((((((((.........)))))))))..)))))...((((((..((((....))))......(((((((.....))))))))))))))))). (-25.14 = -26.10 + 0.96)


| Location | 19,954,362 – 19,954,482 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.92 |
| Mean single sequence MFE | -30.41 |
| Consensus MFE | -21.18 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19954362 120 - 22224390 UCAAUGGAAUCAAAUUCCCAUAUGAAAUUUGGUGUUACUCAAAAGAGGUUGAUUCCCUCAGCGGGAAUUCAAUCAAGUGCAAUGUGAUCAAAUGUUCUGCCACCUUGCCAUUUAAAUUUA .....(((((...)))))......((((((((..............(((((((((((.....))))).))))))..(.((((.(((..............))).)))))..)))))))). ( -23.64) >DroSec_CAF1 66862 120 - 1 UCGACGGAAUCAAAUUCCCAUAUGGAAUUUGGUGUUUCUCGAAAGAGAUUGAUUCCCUCGGCGGGAAUUCAAUCAAGUCUAAUGUGAUCAAAUGUAACGCCACCUUGCCAUUUAAAUAAG ..((.(((((((((((((.....)))))))......((((....))))..)))))).))((((...(((..((((.........))))..)))....))))................... ( -32.00) >DroSim_CAF1 56244 120 - 1 UCGAUGGAAUCAAAUUCCCAUAUGGAAUUUGGUGUUUCUCGAAAGAGAUUGAUUCCCUCGGCGGGAAUUCAAUCAAGUCUAAUGUGAUCAAAUGUAACGCCACCUUGCCAUUUAAAUAAG ..(((((....(((((((.....)))))))((((((.(((....))(((((((((((.....))))).))))))...................).))))))......)))))........ ( -33.30) >DroEre_CAF1 53619 120 - 1 UCGACGGAAUCAAAUUCCCAUAUGGAAUUUUGUGUUUCGCAAAAGAGAUUGAUUCCCACAGCGGGAAUUCUAUCAGGUCUAAUGUGAUCGAGUUUCCCGCCACCUUGUCAAUUAAAUAAG ..(((((((((((.(((((....))..(((((((...)))))))))).))))))))....((((((((((.((((.((...)).)))).))).)))))))......)))........... ( -37.50) >DroYak_CAF1 55969 120 - 1 UCGACCGAAUCAAAUUCCCAUAUGGAAUUCUGUGUUUCUCCAAAGAGAUUGAUUCCCACAGCGGGAAUUCAAUCAAGUCUAAUGUGAUCAAAUGUCCCGCCACCUCGCCACUUAAAUAAG .(((..((((((((((((.....)))))........((((....))))))))))).....(((((((((..((((.........))))..))).))))))....)))............. ( -25.60) >consensus UCGACGGAAUCAAAUUCCCAUAUGGAAUUUGGUGUUUCUCAAAAGAGAUUGAUUCCCUCAGCGGGAAUUCAAUCAAGUCUAAUGUGAUCAAAUGUACCGCCACCUUGCCAUUUAAAUAAG ..(((...((((((((((.....)))))))))).............(((((((((((.....))))).))))))..)))......................................... (-21.18 = -21.98 + 0.80)



| Location | 19,954,402 – 19,954,518 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.99 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -21.54 |
| Energy contribution | -22.90 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19954402 116 + 22224390 GCACUUGAUUGAAUUCCCGCUGAGGGAAUCAACCUCUUUUGAGUAACACCAAAUUUCAUAUGGGAAUUUGAUUCCAUUGAAGAACUUGAGCCUCUAAGCACAAG----CCAACCACAACA ..(((..(..((((((((.....)))))).....))..)..))).........(((((.((((((......)))))))))))..((((.((......)).))))----............ ( -25.10) >DroSec_CAF1 66902 120 + 1 AGACUUGAUUGAAUUCCCGCCGAGGGAAUCAAUCUCUUUCGAGAAACACCAAAUUCCAUAUGGGAAUUUGAUUCCGUCGAAGAACUUGGGCCUCUAAGCACCAGGCAACCAACCACAACA ......((((((.(((((.....)))))))))))((((.(((.......((((((((.....))))))))......))))))).(((((((......)).)))))............... ( -34.02) >DroSim_CAF1 56284 120 + 1 AGACUUGAUUGAAUUCCCGCCGAGGGAAUCAAUCUCUUUCGAGAAACACCAAAUUCCAUAUGGGAAUUUGAUUCCAUCGAAGAACUUGGGCCUCUAAGCACCAGGCAACCAACCACAACA ......((((((.(((((.....)))))))))))((((.(((.......((((((((.....))))))))......))))))).(((((((......)).)))))............... ( -35.12) >DroEre_CAF1 53659 116 + 1 AGACCUGAUAGAAUUCCCGCUGUGGGAAUCAAUCUCUUUUGCGAAACACAAAAUUCCAUAUGGGAAUUUGAUUCCGUCGAAGAACUUGUGCCCCUAAGCCCAAAG----CAACCACAACA .(((..(((.(((((((((.(((((((.........(((((.......))))))))))))))))))))).)))..))).......(((.((......)).)))..----........... ( -24.20) >DroYak_CAF1 56009 116 + 1 AGACUUGAUUGAAUUCCCGCUGUGGGAAUCAAUCUCUUUGGAGAAACACAGAAUUCCAUAUGGGAAUUUGAUUCGGUCGAAGAACUUGGGCCUCUAAGCACAAAG----CAACCACAACA .((((.(((..((((((((.(((((((.((..((((....))))......)).)))))))))))))))..))).))))......(((((....))))).......----........... ( -35.10) >consensus AGACUUGAUUGAAUUCCCGCUGAGGGAAUCAAUCUCUUUCGAGAAACACCAAAUUCCAUAUGGGAAUUUGAUUCCGUCGAAGAACUUGGGCCUCUAAGCACAAGG___CCAACCACAACA ......((((((.(((((.....)))))))))))((((.(((.......((((((((.....))))))))......))))))).((((.((......)).))))................ (-21.54 = -22.90 + 1.36)


| Location | 19,954,402 – 19,954,518 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.99 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -26.94 |
| Energy contribution | -27.98 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19954402 116 - 22224390 UGUUGUGGUUGG----CUUGUGCUUAGAGGCUCAAGUUCUUCAAUGGAAUCAAAUUCCCAUAUGAAAUUUGGUGUUACUCAAAAGAGGUUGAUUCCCUCAGCGGGAAUUCAAUCAAGUGC .((..(((((((----((((.(((....))).)))))...............(((((((..................(((....)))(((((.....))))))))))))))))))...)) ( -29.20) >DroSec_CAF1 66902 120 - 1 UGUUGUGGUUGGUUGCCUGGUGCUUAGAGGCCCAAGUUCUUCGACGGAAUCAAAUUCCCAUAUGGAAUUUGGUGUUUCUCGAAAGAGAUUGAUUCCCUCGGCGGGAAUUCAAUCAAGUCU .....(((((((((.(((((.(((....))))))......((((.(((((((((((((.....)))))))......((((....))))..)))))).)))).)).)).)))))))..... ( -40.10) >DroSim_CAF1 56284 120 - 1 UGUUGUGGUUGGUUGCCUGGUGCUUAGAGGCCCAAGUUCUUCGAUGGAAUCAAAUUCCCAUAUGGAAUUUGGUGUUUCUCGAAAGAGAUUGAUUCCCUCGGCGGGAAUUCAAUCAAGUCU .....(((((((((.(((((.(((....))))))......((((.(((((((((((((.....)))))))......((((....))))..)))))).)))).)).)).)))))))..... ( -40.10) >DroEre_CAF1 53659 116 - 1 UGUUGUGGUUG----CUUUGGGCUUAGGGGCACAAGUUCUUCGACGGAAUCAAAUUCCCAUAUGGAAUUUUGUGUUUCGCAAAAGAGAUUGAUUCCCACAGCGGGAAUUCUAUCAGGUCU ...........----....((((((.(..(((((((((((.....(((((...))))).....)))).)))))))..)........(((.(((((((.....)))))))..))))))))) ( -33.00) >DroYak_CAF1 56009 116 - 1 UGUUGUGGUUG----CUUUGUGCUUAGAGGCCCAAGUUCUUCGACCGAAUCAAAUUCCCAUAUGGAAUUCUGUGUUUCUCCAAAGAGAUUGAUUCCCACAGCGGGAAUUCAAUCAAGUCU .((((.((..(----(((.(.(((....)))).)))).)).)))).(((.((((((((.....)))))).))...)))........(((((((((((.....))))).))))))...... ( -28.80) >consensus UGUUGUGGUUGG___CCUUGUGCUUAGAGGCCCAAGUUCUUCGACGGAAUCAAAUUCCCAUAUGGAAUUUGGUGUUUCUCAAAAGAGAUUGAUUCCCUCAGCGGGAAUUCAAUCAAGUCU .....................((((.((((((((((((((.....(((((...))))).....))))))))).)))))........(((((((((((.....))))).)))))))))).. (-26.94 = -27.98 + 1.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:36 2006