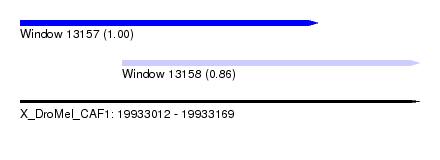
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,933,012 – 19,933,169 |
| Length | 157 |
| Max. P | 0.998919 |

| Location | 19,933,012 – 19,933,129 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -19.90 |
| Energy contribution | -21.03 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.69 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19933012 117 + 22224390 GCCGCGAUUUUUGUGGCAACAUGCGUGCCAACUGGAGGCACAACCACCCAGCUUCCAGCUCCCAGCACCCAGCACCCAGCACCCAGCACCCAGCACCCGGCACCCAACUCAACUCAA (((((((...)))))))....((.(((((...(((((((...........)))))))(((....((.....)).....((.....))....)))....))))).))........... ( -33.70) >DroSec_CAF1 46149 89 + 1 GCCGCGACUUUUGUGGCAACAUGCGUGCCAACUGGAGGCACAACCGCCCAGCUUCCAGCUCCCAG----------------------------CACCCGGCACCCAACUCAACUCAA (((((((...)))))))....((.(((((...(((((((...........)))))))((.....)----------------------------)....))))).))........... ( -30.00) >DroSim_CAF1 38173 89 + 1 GCCGCGACUUUUGUGGCAACAUGCGUGCCAACUGGAGGCACAACCGCCCAGCUUCCAGCUCCCAG----------------------------CACCCGGCACCCAACUCAACUCAA (((((((...)))))))....((.(((((...(((((((...........)))))))((.....)----------------------------)....))))).))........... ( -30.00) >DroYak_CAF1 41542 108 + 1 --CGCGAUUUUUGUGGCAACAUGCGUGCCAACUGGGGGCACAACCACCCAGCUCGCAACACCCAG-------CACACAACACCCAACACCCAGCAUACAACACCCAACUCAACUCAG --.((((......(((((.......))))).(((((((.....)).))))).)))).........-------............................................. ( -21.60) >consensus GCCGCGACUUUUGUGGCAACAUGCGUGCCAACUGGAGGCACAACCACCCAGCUUCCAGCUCCCAG____________________________CACCCGGCACCCAACUCAACUCAA (((((((...)))))))....((.(((((..((((((((...........))))))))(.....).................................))))).))........... (-19.90 = -21.03 + 1.13)


| Location | 19,933,052 – 19,933,169 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.85 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19933052 117 + 22224390 CAACCACCCAGCUUCCAGCUCCCAGCACCCAGCACCCAGCACCCAGCACCCAGCACCCGGCACCCAACUCAACUCAAAUAAAUGCCGGGGCUUUAGUGUUAAUGAGGCAAUGCGUCA ..........(((((..(((....((.....))....)))....(((((..(((.(((((((....................))))))))))...)))))...)))))......... ( -30.85) >DroSec_CAF1 46189 89 + 1 CAACCGCCCAGCUUCCAGCUCCCAG----------------------------CACCCGGCACCCAACUCAACUCAAAUAAAUGCCGGGGCUUUAGUGUUAAUGAGGCAAUGUGUCA .....(((((.......(((...((----------------------------(.(((((((....................))))))))))..))).....)).)))......... ( -23.95) >DroSim_CAF1 38213 89 + 1 CAACCGCCCAGCUUCCAGCUCCCAG----------------------------CACCCGGCACCCAACUCAACUCAAAUAAAUGCCGGGGCUUUAGUGUUAAUGAGGCAAUGCGUCA ....(((...(((((.(((.(..((----------------------------(.(((((((....................))))))))))...).)))...)))))...)))... ( -25.85) >DroYak_CAF1 41580 110 + 1 CAACCACCCAGCUCGCAACACCCAG-------CACACAACACCCAACACCCAGCAUACAACACCCAACUCAACUCAGAUAAAUGCCGGGGCUUUAGUGUUAAUGAGGCAAUGCGUCA .............((((...((((.-------.....(((((......(((.((((.........................)))).)))......)))))..)).))...))))... ( -17.81) >consensus CAACCACCCAGCUUCCAGCUCCCAG____________________________CACCCGGCACCCAACUCAACUCAAAUAAAUGCCGGGGCUUUAGUGUUAAUGAGGCAAUGCGUCA ..........((...........................................(((((((....................)))))))((((((.......))))))...)).... (-14.48 = -14.85 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:20 2006