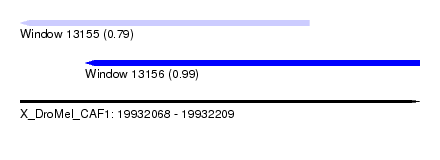
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,932,068 – 19,932,209 |
| Length | 141 |
| Max. P | 0.986692 |

| Location | 19,932,068 – 19,932,170 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -24.67 |
| Energy contribution | -25.37 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
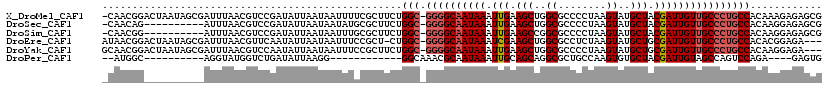
>X_DroMel_CAF1 19932068 102 - 22224390 UUUCGCUUCUGGC-GGGGCAAUAAAUUGAAGCUGGCGCCCCUAAGUAUGCUACGAUUGUUGCCCUGCCACAAAGAGAGCGA-GCAGUCAUAUUAUGCACAUAAA ..(((((((((((-((((((((((.(((.(((..((........))..))).))))))))))))))))....))).)))))-(((.........)))....... ( -38.50) >DroSec_CAF1 45262 102 - 1 AUGCGCUUCUGGC-GGGGCAAUAAAUUGAAGCUGGCGCCCCUAAGUAUGCUACGAUUGUUGCCCUGCCACAAGGAGAGCGA-GCAGUCAUAUUAUGCACAUAAA .(((.((((((((-((((((((((.(((.(((..((........))..))).))))))))))))))))...))))).))).-(((.........)))....... ( -39.00) >DroSim_CAF1 37296 102 - 1 UUGCGCUUCUGGC-GGGGCAAUAAAUUGAAGCCGGCGCCCCUAAGUAUGCUACGAUUGUUGCCCUGCCACAAGGAGAGCGA-GCAGUCAUAUUAUGCACAUAAA ((((.((((((((-((((((((((.(((.(((..((........))..))).))))))))))))))))...))))).))))-(((.........)))....... ( -39.90) >DroEre_CAF1 39094 97 - 1 UUCCGCU-CUGGC-GGGGCAAUAAAUCGAAGCUGGCGCCUCUAAGUAUGCUGCGAUUGUUGCCCUGCCACACGGAGA-----GCAGUCAUAUUAUGCACAUAAA (((((..-.((((-((((((((((.(((.(((..((........))..))).)))))))))))))))))..))))).-----(((.........)))....... ( -40.40) >DroYak_CAF1 40597 98 - 1 UUCCGCUUCUGGC-GGGGCAAUAAAUUGAAGCUGGCGCCCCUAAGUAUGCUGCGAUUGUUGCCCUGCCACAAGGAGA-----GCAGUCAUAUUAUGCACAUAAA ((((.....((((-((((((((((.(((.(((..((........))..))).)))))))))))))))))...)))).-----(((.........)))....... ( -36.00) >DroPer_CAF1 45777 90 - 1 ----------GGCAAACGCAAUAAAUUGCAGCAGGCGCUGCCAAGUGUGCUACGAUUGUAGCCAGUCCAGA----GAGUGACGCACUUAUAUUCUGCACAUAAA ----------.......(((.......(((((....))))).(((((((((((....)))))..((((...----..).)))))))))......)))....... ( -24.40) >consensus UUCCGCUUCUGGC_GGGGCAAUAAAUUGAAGCUGGCGCCCCUAAGUAUGCUACGAUUGUUGCCCUGCCACAAGGAGAGCGA_GCAGUCAUAUUAUGCACAUAAA ..........(((.((((((((((.(((.(((..((........))..))).))))))))))))))))..............(((.........)))....... (-24.67 = -25.37 + 0.69)



| Location | 19,932,091 – 19,932,209 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.03 |
| Mean single sequence MFE | -37.15 |
| Consensus MFE | -22.87 |
| Energy contribution | -23.57 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19932091 118 - 22224390 -CAACGGACUAAUAGCGAUUUAACGUCCGAUAUUAAUAAUUUUCGCUUCUGGC-GGGGCAAUAAAUUGAAGCUGGCGCCCCUAAGUAUGCUACGAUUGUUGCCCUGCCACAAAGAGAGCG -...(((((...............)))))..............((((((((((-((((((((((.(((.(((..((........))..))).))))))))))))))))....))).)))) ( -39.96) >DroSec_CAF1 45285 108 - 1 -CAACAG----------AUUUAACGUCCGAUAUUAAUAAUAUGCGCUUCUGGC-GGGGCAAUAAAUUGAAGCUGGCGCCCCUAAGUAUGCUACGAUUGUUGCCCUGCCACAAGGAGAGCG -......----------.........................((.((((((((-((((((((((.(((.(((..((........))..))).))))))))))))))))...))))).)). ( -36.60) >DroSim_CAF1 37319 108 - 1 -CAACGG----------AUUUAACGUCCGAUAUUAAUAAUUUGCGCUUCUGGC-GGGGCAAUAAAUUGAAGCCGGCGCCCCUAAGUAUGCUACGAUUGUUGCCCUGCCACAAGGAGAGCG -...(((----------((.....))))).............((.((((((((-((((((((((.(((.(((..((........))..))).))))))))))))))))...))))).)). ( -42.10) >DroEre_CAF1 39116 115 - 1 AUAACGGACUAAUAGCGAUUUAACGUUCAAUAUUAAUAAUUUCCGCU-CUGGC-GGGGCAAUAAAUCGAAGCUGGCGCCUCUAAGUAUGCUGCGAUUGUUGCCCUGCCACACGGAGA--- .........(((((..((........))..)))))....((((((..-.((((-((((((((((.(((.(((..((........))..))).)))))))))))))))))..))))))--- ( -40.90) >DroYak_CAF1 40619 116 - 1 GCAACGGACUAAUAGCGAUUUAACGUCCAAUAUUAAUAAUUUCCGCUUCUGGC-GGGGCAAUAAAUUGAAGCUGGCGCCCCUAAGUAUGCUGCGAUUGUUGCCCUGCCACAAGGAGA--- .....((((...............))))...........(((((.....((((-((((((((((.(((.(((..((........))..))).)))))))))))))))))...)))))--- ( -39.06) >DroPer_CAF1 45801 92 - 1 --AUGGC----------AGGUAUGGUCUGAUAUUAAGG------------GGCAAACGCAAUAAAUUGCAGCAGGCGCUGCCAAGUGUGCUACGAUUGUAGCCAGUCCAGA----GAGUG --.((((----------((((.((((((.........)------------)))....((((....))))..)).)).))))))..((.(((((....))))))).......----..... ( -24.30) >consensus _CAACGG__________AUUUAACGUCCGAUAUUAAUAAUUUCCGCUUCUGGC_GGGGCAAUAAAUUGAAGCUGGCGCCCCUAAGUAUGCUACGAUUGUUGCCCUGCCACAAGGAGAGCG ..................................................(((.((((((((((.(((.(((..((........))..))).))))))))))))))))............ (-22.87 = -23.57 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:18 2006