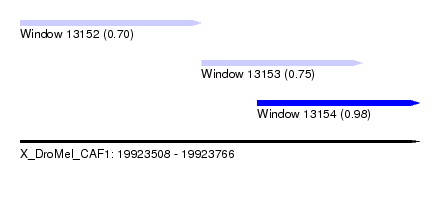
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,923,508 – 19,923,766 |
| Length | 258 |
| Max. P | 0.981910 |

| Location | 19,923,508 – 19,923,625 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.14 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -20.04 |
| Energy contribution | -21.64 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19923508 117 + 22224390 CAUGCUGCAAAGUCA-GUUCUGCGAGCUUACUUCCAACUUUUUGCGUAUUUCAUAAUUUGUG--UGUGCUGCAUAUUUCACAUAUUCAUGAAAUUCCGAGAAUAAGCGCAGUACAUGAUG ..((((((.((((.(-((((...))))).)))).....((((((.(.(((((((....((((--((............))))))...))))))).))))))).....))))))....... ( -26.90) >DroSec_CAF1 37677 117 + 1 CAUGCUGCAAAGUCA-GUUCUGCGAGUUUGCUCGCAACUUUUUUCGUAUUUCAUAAUUUGUG--UGUGUUGCAUAUUUCACAUAUUCAUGAAAUUCCGAGAACAAGCGCAGUACAUGAUG ..((((((.......-....((((((....)))))).(((((((((.(((((((.....(((--((((..........)))))))..)))))))..)))))).))).))))))....... ( -33.60) >DroSim_CAF1 29118 117 + 1 CAUGCUGCAAAGUCA-GUUCUGCGAGUUUGCUCGCAACUUUUUUCGUAUUUCAUAAUUUGUG--UGUGUUGCAUAUUUCACAUAUUCAUGAAAUUCCGAGAACAAGCGCAGUACAUGAUG ..((((((.......-....((((((....)))))).(((((((((.(((((((.....(((--((((..........)))))))..)))))))..)))))).))).))))))....... ( -33.60) >DroEre_CAF1 34683 105 + 1 CAUGUUGCAUAGUCA-GUUCGCCGGC------------UUUUUUUGUAUUUCGUAAUUUGUG--UGUGUUGCAUAUUUCACAUAUUCAUGAAAUUCCGAGAAUAAGCUCGCUACAUGAUG ...........((((-....((.(((------------((((((((.(((((((.....(((--((((..........)))))))..)))))))..)))))).))))).))....)))). ( -24.30) >DroYak_CAF1 36098 120 + 1 CAUGUUGCAAAGUCAUGUUCUCCGACUUUGCUCGCAACUUUUUUUGUAUUUCAUAAUUUGUGUGUGUGUUGCAUAAUUCACAUAUUCAUGAAAUUCUGAGAAUAAGCGCACUAUAUGAUG (((((.((((((((.........)))))))).(((....(((((...((((((((((.((((..((((...))))...)))).))).)))))))...)))))...)))....)))))... ( -28.40) >consensus CAUGCUGCAAAGUCA_GUUCUGCGAGUUUGCUCGCAACUUUUUUCGUAUUUCAUAAUUUGUG__UGUGUUGCAUAUUUCACAUAUUCAUGAAAUUCCGAGAAUAAGCGCAGUACAUGAUG ((((((((............((((((....))))))....((((((.((((((((((.((((..((((...))))...)))).))).)))))))..)))))).....))))..))))... (-20.04 = -21.64 + 1.60)


| Location | 19,923,625 – 19,923,729 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.99 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -30.70 |
| Energy contribution | -30.18 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19923625 104 + 22224390 UACC----CGGCACUUCCACUCACAUCCCUGUGACAAUGUACGUUGCACACGGCAACGGC------------GGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGUUGCCGUCAUAAGC ....----..((........(((((....)))))........(((((.....)))))(((------------((((((((.((((((((....)).)).)))))))))))))))....)) ( -33.80) >DroSec_CAF1 37794 104 + 1 UACC----CGGCACUGCCACUCGCAUCCCUGUGACAAUGCACGUUGCACACGGCAACGGC------------GGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGCUGCCGUCAUAAGC ....----.(((...)))...........((((((...((.((((((.....))))))))------------((((((((.((((((((....)).)).))))))))))))))))))... ( -40.90) >DroSim_CAF1 29235 104 + 1 UACC----CGGCACUGCCACUCGCAUCCCUGUGACAAUGCACGUUGCACACGGCAACGGC------------GGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGCUGCCGUCAUAAGC ....----.(((...)))...........((((((...((.((((((.....))))))))------------((((((((.((((((((....)).)).))))))))))))))))))... ( -40.90) >DroEre_CAF1 34788 102 + 1 UACCC------CAGUACCACUCACAUCCCUGUGACAAUGCACGCUGCACACGGCAACGGC------------GGCGGCAUUAGUGACCUAGUUUGAGUCCGCUGUAUUGCCGUCAUAAGC .....------..(((....(((((....)))))...)))..(((....(((((((..((------------(((((.(((...(((...)))..)))))))))).)))))))....))) ( -30.80) >DroYak_CAF1 36218 120 + 1 CACCUACCCGGCACUGCCACUCACAUCCCUGUGACAAUGCACGUUGCACGUGGCAACGGCAACGGUGGCGUCGGUGGCAUUAGUGACCUAGUUUGUGUCCGCUGUGUUGCCGCCAUAAGC .........(((...((((((((((....))))).......((((((.(((....))))))))))))))...((..((((.(((((((......).)).))))))))..)))))...... ( -47.00) >consensus UACC____CGGCACUGCCACUCACAUCCCUGUGACAAUGCACGUUGCACACGGCAACGGC____________GGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGUUGCCGUCAUAAGC ....................(((((....)))))....((.((((((.....))))))))............(((((((.((((((((......).)).)))))...)))))))...... (-30.70 = -30.18 + -0.52)



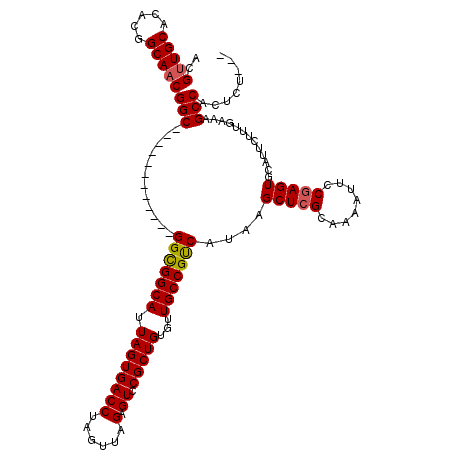
| Location | 19,923,661 – 19,923,766 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.64 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -32.24 |
| Energy contribution | -32.32 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19923661 105 + 22224390 ACGUUGCACACGGCAACGGC------------GGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGUUGCCGUCAUAAGCUCGCAAAAUUCCGAGUACAUUCUUUGAAAGCCACUCA--- ..(((((.....)))))(((------------(((((((.(((((((((....)).)).)))))...)))))))....(((((........))))).............))).....--- ( -36.20) >DroSec_CAF1 37830 105 + 1 ACGUUGCACACGGCAACGGC------------GGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGCUGCCGUCAUAAGCUCGCAAAAUUCCGAGUGCAUUCUUUGAAAGCCACUCU--- ..(((((.....)))))(((------------((((((((.((((((((....)).)).)))))))))))))))..................((((((.(((...))).).))))).--- ( -38.80) >DroSim_CAF1 29271 105 + 1 ACGUUGCACACGGCAACGGC------------GGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGCUGCCGUCAUAAGCUCGCAAAAUUCCGAGUGCAUUCUUUGAAAGCCACUCU--- ..(((((.....)))))(((------------((((((((.((((((((....)).)).)))))))))))))))..................((((((.(((...))).).))))).--- ( -38.80) >DroEre_CAF1 34822 108 + 1 ACGCUGCACACGGCAACGGC------------GGCGGCAUUAGUGACCUAGUUUGAGUCCGCUGUAUUGCCGUCAUAAGCUGGCAAAAUUCCGAGUGCAUUCUUUGAAAGCCACUCCCCA ..(((((...((....))..------------(((((((.(((((..((......))..)))))...)))))))....)).)))........((((((.(((...))).).))))).... ( -34.10) >DroYak_CAF1 36258 117 + 1 ACGUUGCACGUGGCAACGGCAACGGUGGCGUCGGUGGCAUUAGUGACCUAGUUUGUGUCCGCUGUGUUGCCGCCAUAAGCUCGCAAAAUUCCGAGUGCAUUCUUUGAAAGCCACUCU--- .((((((.(((....)))))))))(((((...(((((((.((((((((......).)).)))))...)))))))....(((((........))))).............)))))...--- ( -46.40) >consensus ACGUUGCACACGGCAACGGC____________GGCGGCAUUAGUGACCUAGUUAGAGUCCGCUGUGUUGCCGUCAUAAGCUCGCAAAAUUCCGAGUGCAUUCUUUGAAAGCCACUCU___ ..(((((.....)))))(((............(((((((.((((((((......).)).)))))...)))))))....(((((........))))).............)))........ (-32.24 = -32.32 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:16 2006